Referring to SARS-CoV-2 Proteins in BEL
Many of the proteins in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
are cleavage products of the replicase polyprotein 1ab (uniprot:P0DTD1).
Unfortunately, the bioinformatics community is not so comfortable with proteins
like this and nomenclature remains tricky. Luckily, the Biological Expression Language (BEL)
has exactly the right tool to encode information about these proteins using the fragment() function.
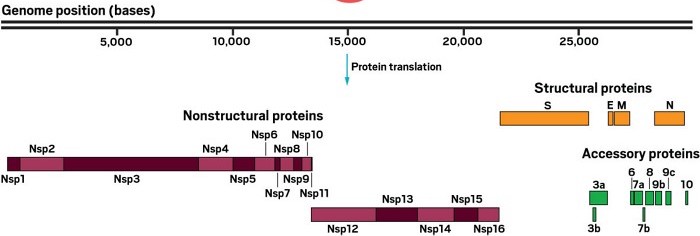
This image was modified from the C&EN article What do we know about the novel coronavirus’s 29 proteins?
UniProt lists each of the 16 non-structural proteins (often written as symbols nsp1-nsp16) as protein chains
of the main protein entry, uniprot:P0DTD1. These chains are assigned identifiers
following the regular expression pattern of PRO_\d{10}. The Identifiers.org registered this pattern
under the prefix uniprot.chain. While it resolves
to URLs following the pattern of https://www.uniprot.org/uniprot/<uniprot_id>#<chain_id>, it appears that the
parent protein’s UniProt identifier is looked up automatically . This is really good news and means that we can
start using stable CURIEs to identify these proteins, even if like me, you’ve never used this prefix before.
Alternatively, BEL allows you to write out the relationship between the parent protein and the
fragment using the fragment() / frag() function (docs).
For example, the nsp1 fragment from position 1-180 can be written in BEL either as
p(uniprot.chain:PRO_0000449619) or as a fragment p(uniprot:P0DTD1 ! R1AB_SARS2, frag(1_180)).
The entire table of non-structural proteins is written out below for your copy/paste convenience
in BEL coding.
| Symbol | Chain | Positions | Name | BEL |
|---|---|---|---|---|
| nsp1 | PRO_0000449619 | 1 – 180 | Host translation inhibitor nsp1 | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(1_180)) |
| nsp2 | PRO_0000449620 | 181 – 818 | Non-structural protein 2 | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(181_818)) |
| nsp3 | PRO_0000449621 | 819 – 2763 | Non-structural protein 3 | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(819_2763)) |
| nsp4 | PRO_0000449622 | 2764 – 3263 | Non-structural protein 4 | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(2764_3263)) |
| nsp5 | PRO_0000449623 | 3264 – 3569 | 3C-like proteinase | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(3264_3569)) |
| nsp6 | PRO_0000449624 | 3570 – 3859 | Non-structural protein 6 | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(3570_3859)) |
| nsp7 | PRO_0000449625 | 3860 – 3942 | Non-structural protein 7 | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(3860_3942)) |
| nsp8 | PRO_0000449626 | 3943 – 4140 | Non-structural protein 8 | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(3943_4140)) |
| nsp9 | PRO_0000449627 | 4141 – 4253 | Non-structural protein 9 | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(4141_4253)) |
| nsp10 | PRO_0000449628 | 4254 – 4392 | Non-structural protein 10 | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(4254_4392)) |
| nsp12 | PRO_0000449629 | 4393 – 5324 | RNA-directed RNA polymerase | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(4393_5324)) |
| nsp13 | PRO_0000449630 | 5325 – 5925 | Helicase | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(5325_5925)) |
| nsp14 | PRO_0000449631 | 5926 – 6452 | Proofreading exoribonuclease | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(5926_6452)) |
| nsp15 | PRO_0000449632 | 6453 – 6798 | Uridylate-specific endoribonuclease | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(6453_6798)) |
| nsp16 | PRO_0000449633 | 6799 – 7096 | 2’-O-methyltransferase | p(uniprot:P0DTD1 ! R1AB_SARS2, frag(6799_7096)) |
I’m not sure what happened to #11. UniProt isn’t listing it here. There’s also the Replicase polyprotein 1a, which lists nsp1-nsp11, but I’m not sure what the difference is yet.
When I first started writing this, I wasn’t actually aware of the existence of the uniprot.chain entry
in Identifiers.org. This makes things a lot better! However, this leaves two tasks for me:
- Integrate the
uniprot.chainnomenclature into PyOBO such that identifiers can be validated and easily resolved to names -
Generate equivalence relationships in BEL linking the CURIE-named and ontologically-defined versions of each as in:
p(uniprot.chain:PRO_0000449619) equivalentTo p(uniprot:P0DTD1 ! R1AB_SARS2, frag(1_180)) ... p(uniprot.chain:PRO_0000449633) equivalentTo p(uniprot:P0DTD1 ! R1AB_SARS2, frag(6799_7096))
Happy BEL coding!