Research
My research is focused on the generation and application of biological knowledge graphs in drug discovery and precision medicine. Recently, I’ve had a specific focus on knowledge graph embedding methodologies and downstream application of link prediction.
Biocuration
While there are several useful public terminologies useful for curation of biomedical relations, there is often the need to develop new controlled vocabularies, thesauri, taxonomies, and ontologies to support new biological phenomena. I lead the team that created the Curation of Neurodegeneration Supporting Ontology (CONSO).
After identifying named entities within scholarly articles, their relations can be extracted and encoded in a knowledge graph. I lead the team that created the knowledge graph Curation of Neurodegeneration in BEL (CONIB) and later the knowledge graph TauBase.
I also lead the same team to re-curate the knowledge graphs curated and published during the AETIONOMY project and developed a novel rational enrichment workflow. Because of the time and cost of curation, prioritization of articles is crucial. I’ve developed semi-automated curation workflows based on a new metric for information density in regions of knowledge graphs.
I also serve on the Biological Expression Language Committee that facilitates the improvement of the language as a modeling paradigm in systems and networks biology.
Data Integration and Harmonization
In order to check the syntax and semantics of these knowledge graphs, I developed PyBEL. To interactively explore these graphs in a web-based environment and identify biological contractions, I developed BEL Commons.
Finally, to integrate all of the rich biological data sources available to the public, I developed Bio2BEL. During the process, I was able to support the ComPath project, which used the Bio2BEL framework to support the curation of equivalencies and hierarchical relations between entries in several major pathway databases (e.g., KEGG, WikiPathways, Reactome) and then later in the PathMe project where they were harmonized in BEL as a common schema.
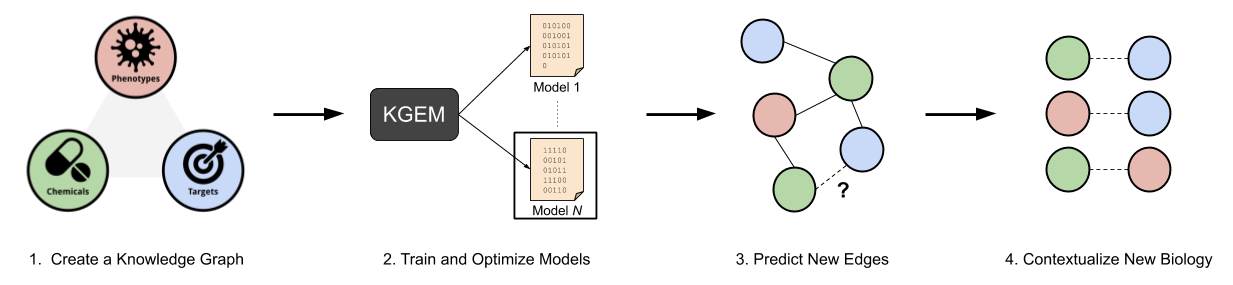
Knowledge Graph Embeddings
Knowledge graph embedding methods learn latent representations for the nodes and edges in a graph to support clustering, link prediction, entity disambiguation, and other downstream machine learning tasks.
I’ve worked on PyKEEN, a PyTorch reimplementation of several recent knowledge graph embedding models with a focus on reproducibility. I’ve also developed BioKEEN, which connects biological knowledge graphs in BEL (notably from Bio2BEL) directly to the PyKEEN pipeline.
Predictions
The link prediction task in knowledge graphs is isomorphic to several tasks in drug discovery and precision medicine.
Predicting links between genes/proteins and diseases accomplishes target identification/prioritization. I’ve worked on GuiltyTargets, which embedded proteins from protein-protein interaction networks annotated with disease-specific differential gene expression patterns. These embeddings were used for positive-unlabeled learning using disease-specific gene lists. While this method works well, it was only single-task (only working on one disease at a time).
Predicting links between chemicals and diseases accomplishes drug repositioning (in the case when the chemical is a known drug) or otherwise novel drug discovery. I’ve worked on DrugReLink, which uses Hetionet to make these predictions for a given chemical or disease.
Because many compounds fail in the clinic due to undesirable side effects, predicting them during early-stage drug discovery could drastically improve the efficiency. I’ve worked on SEffNet, which uses a network composed of drug-disease, drug-side effect, drug-target, and drug-drug links to predict compounds’ side effects and give insight into the targets mediating those side effects.
Some of my ongoing work is to apply these methods in precision medicine. I’m doing it by annotating patients as nodes in networks, and creating edges to biological entities based on clinical measurements (e.g., gene expression) then embedding those nodes for downstream machine learning tasks such as subgroup identification and survival analysis.
Presentations
- Bioregistry Workshop at Biocuration 2025 at Bioregistry Workshop @ Biocuration 2025
- Bioregistry and the NFDI in 2024 at 3rd Ontologies4Chem Workshop
- Modern software development practice with Python (invited) at May Institute
- Assembly and inference over semantic mappings to support the NFDI Terminology Service (invited) at TS4NFDI Community Workshop
- Assembly and Reasoning over Semantic Mappings at Scale at Biocuration 2024
- Assembly of Domain Knowledge at Scale in Biomedicine and Beyond (invited) at Harvard Medical School - Laboratory of Systems Pharmacology Meeting
- Machine-assisted integration of data and knowledge at scale to support biomedical discovery (invited) at NIH BISTI Seminar
- Introduction to WPCI 2023 at Winter 2023 Workshop on Prefixes, CURIEs, and IRIs
- Democratizing Biocuration, or, How I Learned to Love the Drive-by Curation (invited) at International Society of Biocuration Annual General Meeting
- Standardization of chemical prefixes, CURIEs, URIs, and semantic mappings at Ontologies4Chem Workshop 2023
- Improving the reproducibility of cheminformatics workflows with chembl-downloader at RDKit User Group Meeting 2023
- Improving ontology interoperability with Biomappings (invited) at OBO Academy - Monarch Training Series
- Modern prefix management with the Bioregistry and `curies` (invited) at OBO Academy - Monarch Training Series
- Promoting the longevity of curated scientific resources through open code, open data, and public infrastructure at Biocuration 2023
- Using dashboards to monitor ontology standardisation and community activity at Ontology Summit 2023
- Introduction to WPCI 2022 at 2022 Workshop on Prefixes, CURIEs, and IRIs
- The Bioregistry, CURIEs, and OBO Community Health at International Conference on Biomedical Ontology (ICBO)
- Axiomatizing Chemical Roles (invited) at Ontologies4Chem Workshop 2022
- Closing the Semantic Gap: Identifying Missing Mappings and Merging Equivalent Concepts to Support Knowledge Graph Assembly at Harvard Medical School - Sorger Lab Meeting
- Modern Scientific Software Development Practice in Python (invited) at May Institute
- Knowledge Graph Embedding with PyKEEN in 2022 (invited) at Knowledge Graph Conference (KGC 2022)
- A Unified Framework for Rank-based Evaluation Metrics for Link Prediction in Knowledge Graphs at Graph Learning Benchmarks (GLB 2022)
- The Biopragmatics Stack: Biomedical and Chemical Semantics for Humans (invited) at Machine-Actionable Data Interoperability for Chemical Sciences (MADICES)
- Reusable Science in Python (invited) at May Institute
- Introduction to WPCI 2021 at 2021 Workshop on Prefixes, CURIEs, and IRIs
- Biomappings: Community Curation of Mappings between Biomedical Entities (poster) at 4th Session of the International Society of Biocuration 2021 Virtual Conference
- Current Issues in Theory, Reproducibility, and Utility of Graph Machine Learning in the Life Sciences (invited) at Graph Machine Learning in Industry
- The Bioregistry: A Metaregistry for Biomedical Entities at 12th International Conference on Biomedical Ontologies
- Perspectives on Knowledge Graph Embedding Models in/out of Biomedicine (invited) at AstraZeneca
- Future Directions for WikiPathway Meta-curation at WikiPathways Developers Conference Call
- The Biological Expression Language and PyBEL in 2020 at COVID-19 Disease Map Community Meeting
- Introduction to the Biological Expression Language and the Rational Enrichment Workflow (invited) at CoronaWhy
- Applications of Knowledge Graphs in Drug Discovery (invited) at Computational Drug Discovery Group, University of Leiden
- Maintenance and Enrichment of Disease Maps in Biological Expression Language (poster) at 4th Disease Maps Community Meeting
- Generation and Application of Biomedical Knowledge Graphs (invited) at Harvard Medical School
- Identifying Drug Repositioning Candidates using Representation Learning on Heterogeneous Networks (poster) at The Eighth Joint Sheffield Conference on Chemoinformatics
- The PyBEL Ecosystem in 2018 at OpenBEL Community Meeting
- From Knowledge Assembly to Hypothesis Generation at Bio-IT World
- Knowledge Assembly in Systems and Networks Biology (poster) at Bio-IT World
- The Human Brain Pharmacome: An Overview (poster) at 3rd European Conference on Translational Bioinformatics
- Gene Set Analysis using Phenotypic Screening Data (poster) at Research, Innovation and Scholarship Expo 2015
Publications
My publications are listed in many places, so there’s not much good in writing it all again on this site. Check one of the following: