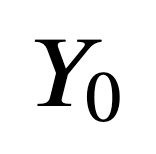I’ve created many open-source projects as well as made significant contributions to others:
 Bio2BEL
Bio2BEL

A framework for reproducible data integration in BEL and knowledge graph construction
 BEL Commons
BEL Commons

A web application for the interactive exploration of networks encoded in BEL
Bioontologies

Access and processing of ontologies on top of ROBOT and OBO Graphs
 PyOBO
PyOBO

Harmonization of biological ontologies and controlled vocabularies
curies

Idiomatic conversion between URIs and compact URIs (CURIEs)
 GuiltyTargets
GuiltyTargets

Target prioritization framework using gene expression and network representation learning
 RatVec
RatVec

Sequence-based representation learning
 PS4DR
PS4DR

Drug repositioning based on bioactivity pattern matching and GWAS
 SeffNet
SeffNet

Drug repositioning framework based on network representation learning
 CLEP
CLEP

Patient stratification framework based on network representation learning
 BEL2SCM
BEL2SCM

Generation of structural causal models (SCMs) from BEL
 STonKGs
STonKGs

Multimodal Transformers for biomedical text and Knowledge Graph data
 ChemicalX
ChemicalX

A deep learning library for drug-drug interaction, polypharmacy side effect, and synergy prediction.
 PathMe
PathMe

Web application for exploration of pathway databases
 Pathway Forte
Pathway Forte

Benchmarking pathway databases in functional enrichment analysis and prediction methods
 INDRA
INDRA

Automated knowledge assembly and modeling in biomedicine
INDRA CoGEx

A $10^8$ relation-scale knowledge graph extending on causal knowledge from INDRA
MIRA

Machine-assisted scientific modeling using meta-model templates and domain knowledge graphs
Gilda

Biomedical named entity recognition and grounding using machine-learned disambiguation
Databases
I’ve created many databases myself through curation, automated assembly, and also by coordinating
others.
 Bioregistry
creator, maintainer
Bioregistry
creator, maintainer

An integrative meta-registry of biological databases, ontologies, and nomenclatures.
 Biomappings
creator, maintainer
Biomappings
creator, maintainer

Predicted and curated mappings between named biological entities
 Biolookup
creator, maintainer
Biolookup
creator, maintainer

Comprehensive database of identifiers, names, synonyms, cross-references, properties, and relations for biomedical entities
 Bioversions
creator, maintainer
Bioversions
creator, maintainer

Automated tracking of the current version for each biological database?
 CONSO
creator, maintainer
CONSO
creator, maintainer

Ontology of phenomena related to neurodegeneration
 CONIB
creator, maintainer
CONIB
creator, maintainer

Curated knowledge graphs describing neurodegeneration in BEL
 ComPath
contributor
ComPath
contributor

A data integration pipeline, manually curated dataset of mappings between pathway databases, and web application for integrative analysis of pathways
 GuiltyTargets
GuiltyTargets
 ChemicalX
ChemicalX
 Bioregistry
creator, maintainer
Bioregistry
creator, maintainer
 Bioversions
creator, maintainer
Bioversions
creator, maintainer