Encoding Biology in Knowledge Graphs
How many molecular biology papers have you read today? This week? This month? If you’re like me, its not so many, and we’re falling behind very quickly. Here’s a chart made by the new PubMed that summarizes how many papers were published mentioning RAS in the last 50 years.
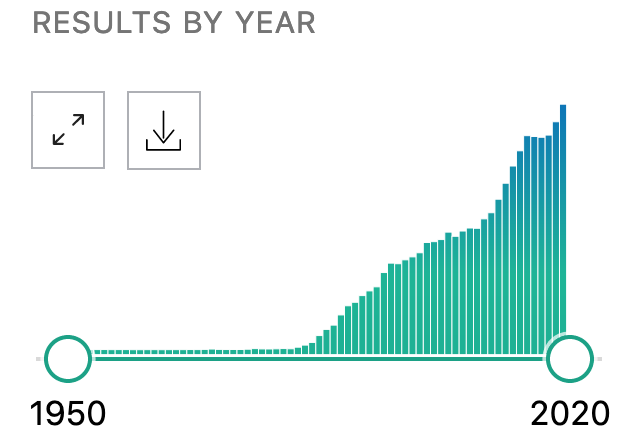
There were 4,483 publications listed in 2019. We can’t read that much, and even if we did, we couldn’t remember it all. That’s why we need to take the knowledge out of the unstructured text and store it in a structured form that can be read and stored in computers. This way, we can easily share it, query it, and write algorithms that can help us reason about the incredible amount of biological knowledge out there.
There are several formats in which this kind of information can be stored on a continuum between directly representing mechanistic biology to representing the knowledge itself. In the popular middle ground are BioPAX and BEL, which I’ll come back to in future posts.
It’s important to keep in mind that knowledge needs to be curated - this can either be manual, through natural language processing, or a mixture of both. I’ve written a paper on such a process, but for now this post should motivate a few following ones describing what it takes to deal with nomenclature, build ontologies, and then start extracting mechanistic biology from the literature.