The Curation of Neurodegeneration Supporting Ontology
While I led the curation program in the Human Brain Pharmacome project during my Ph.D. from 2018-2019 at Fraunhofer, we built the Curation of Neurodegeneration Supporting Ontology (CONSO). This post outlines the project’s needs for quality control and re-curation that lead to its generation, the curation process, and how CONSO constitutes an example of how to follow the guidelines I proposed in a previous blog post on building ontologies.
Before I joined Fraunhofer for my master’s and Ph.D., previous group members had made first attempts at generating several narrow ontologies, terminologies, and controlled vocabularies within the field of neurodegenerative disease research which were then used as controlled vocabularies (CVs) in knowledge graphs curated for the AETIONOMY project. However, the project did not outline a maintenance plan and around late 2015, AETIONOMY’s curation program effectively ended.
Three years later (at the start of 2018), I was tasked with revitalizing the curation program as part of the Human Brain Pharmacome project. This meant assessing the quality and currentness of knowledge graphs and their underlying CVs before leading a new curation campaign to enrich them (to be described in a future blog post). The remainder of this blog post is about the issues with these underlying ontologies, how we addressed them, and a description of the resulting ontology that we generated.
Assessing Controlled Vocabularies used by NeuroMMSig
The first step was to categorize the ontologies, terminologies, and CVs that were used in the knowledge graphs (the set of which comprises NeuroMMSig (paper, website) from now on) into high and low confidence. The first criterion for high confidence was that they were either in a repository of high quality nomenclatures (either OLS or OBO Foundry) or they were registered in the MIRIAM database at Identifiers.org. The second criterion for high confidence was that they were being actively maintained in a version controlled repository like the Human Phenotype Ontology or FamPlex. Because there are a few examples of standard ontologies that are not maintained, the confidence labels help convey this distinction. As an aside, the difference between ontology, terminology, and CV is not important for their application in the curation of knowledge graphs (see my previous blog post). For the remainder of this post, I’ll be calling them CVs.
The list of CVs used in NeuroMMSig that did not meet the high confidence criteria included three previously published ontologies (i.e., ADO, PDON, MSO) that could be found in the Protégé OWL format through the NCBO BioPortal. The underlying issue with these three resources is that they are actually text mining dictionaries masquerading as ontologies. As outlined in their respective publications, they were created to train a dictionary-based named entity recognition system. From there, careless usage of Protégé led to confusing organization, messy and cryptic entity labels, lack of meaningful entity hierarchies, lack of definitions or provenance for each entity, lack of curator provenance for each entity, and non-compliance with the MIRIAM standards for entity identifiers. Further, the lack of maintenance meant that when entities originally novel to these resources were curated again for inclusion in high confidence, mainstream ontologies, that there would exist no mappings (e.g., equivalences) to these high confidence resources. This could create redundancies that undermine the value of the downstream resources, like NeuroMMSig, that use these low confidence resources. Like I outlined in my previous post, the solution to this problem is to not compete with groups maintaining high confidence resources, and instead to contribute to them directly.
A fourth previously published CV (i.e., NIFT) in NeuroMMSig that did not meet the high confidence criteria only existed as a Biological Expression Language (BEL) namespace file served by Fraunhofer’s web-based BEL curation interface. Because it could be shut down at any time, I started by copying it to a more persistent server but later began using GitHub directly for versioning and distribution. While not masquerading as an ontology, this CV also had issues with messy names (inclusion of strange punctuation, inconsistent usage of capitalization, etc.). By virtue of its lack of structure or organization that benefits ontologies, the assessment of each term for its novelty, correctness, and whether it could be replaced with an already existing entry in a high confidence ontology became its own time-consuming research project.
There were additional low confidence CVs generated as ad-hoc extensions to high confidence CVs. For example, the Brain Region and Cell-type Ontology (BRCO; unpublished) was an extension to Uberon. Unfortunately, the value of this extension was questionable as it seemed most (if not all) of the extensions made actually duplicated efforts of the original ontology itself. As several years had passed between the generation of this extension and my assessment, it’s possible that some of the terms included in BRCO were later curated by Uberon by virtue of its excellent maintenance. This phenomenon persisted across other examples of ad-hoc ontology extensions I found in NeuroMMSig.
Finally, there were several other novel low confidence CVs only distributed as BEL namespaces that suffered from a mixture of the previously described problems.
Making a Home for Lost and Forgotten Names
It should also be noted that my assessment of the low confidence CVs focused on their respective terms that appeared in the NeuroMMSig knowledge graphs. There were also hundreds of terms appearing in NeuroMMSig that had not been assigned based on any CV at all (what I called “naked names”). Some could be assigned via text mining tools, but others needed a home. It was finally time to start curating an ontology that could unify all of the existing abandoned CVs used by NeuroMMsig and solve their respective problems. And so, along with my team of excellent students, work was begun on the Curation of Neurodegeneration Supporting Ontology (CONSO; data, website).
We began by re-curating all of the terms from low confidence CVs that appeared in NeuroMMSig. Each was given its own entry in CONSO along with a persistent identifier, a typographically sane preferred label, a definition in english prose, references to papers where it appeared, and contact information of the curator in case there were questions. We found that the curator attribution was incredibly important as questions arose about previous terms. We maintained synonyms and xrefs for each term as well as implementing code that leverages several services (e.g., OLS, PubChem, etc.) for new mappings to propose for curation. After, we did the same for all of the names appearing in NeuroMMSig that were not qualified with a namespace/prefix. Later, CONSO was improved with completely novel curation to support additional curation of neurodegeneration disease phenomena that became the Curation of Neurodegeneration in BEL (CONIB; data, website).
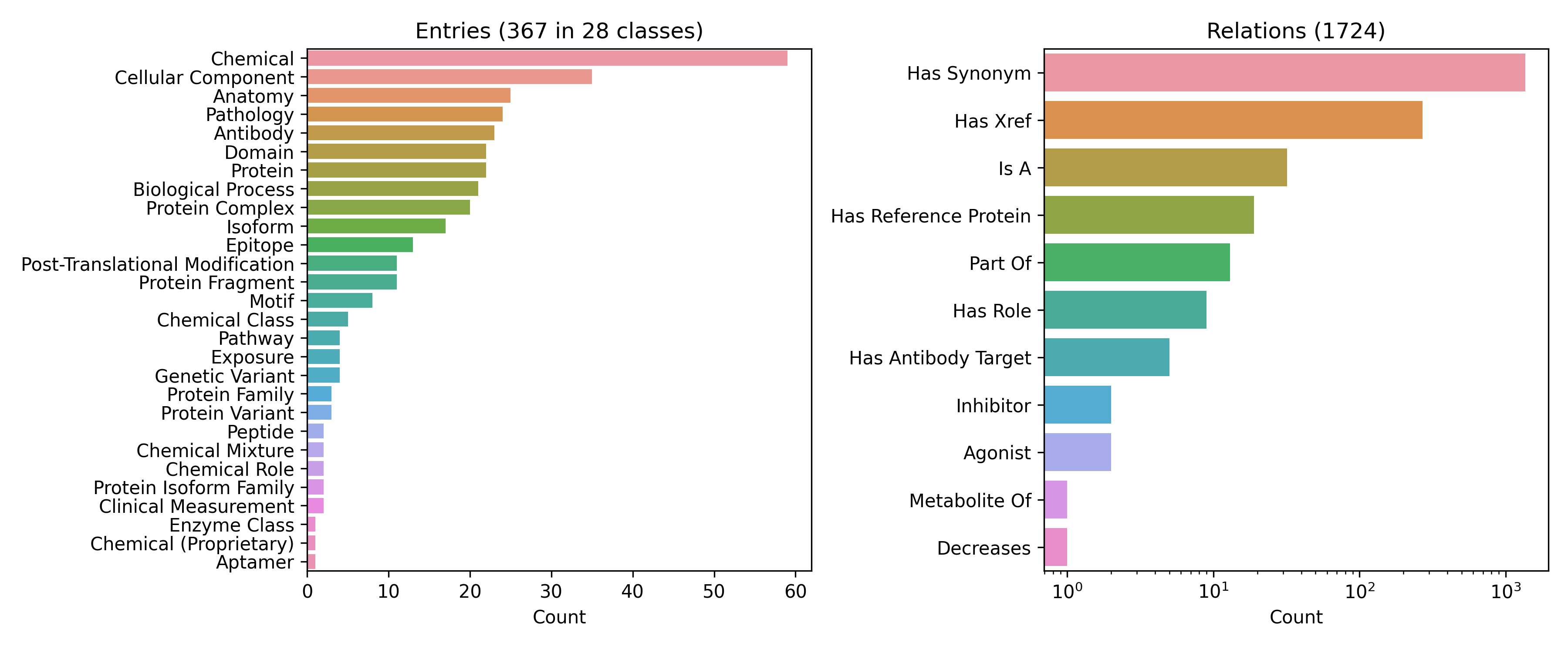
CONSO maintains entities of several types, including chemicals, chemical roles, proteins, protein families, protein variants, protein isoforms, and protein isoform families (see figure below). It also maintains its own vocabulary of relationships and instances of those relationships. For example, we were unable to find a relation that describes the epitope corresponding to a given antibody in a high quality resource (e.g., the Relation Ontology). Therefore, we created our own relationship and started curating instances of it. Because it’s going as far as defining its own semantics, CONSO is a proper ontology.
While CONSO’s hierarchy is currently neither aligned to the Basic Formal Ontology nor the Systems Biology Ontology for the classification of entity types, the current organizational scheme is best suited to support downstream curation of biomedical relations. While I’ve written quite a bit about the applications of terminologies and ontologies by this point, I am definitely not an expert in its pedagogy and will have to bring in others for discussion on how to do this best. One of the other reasons why I haven’t done that yet is that we are slowly making pull requests, issues, and contributions to other ontologies to move all of the best stuff out of CONSO into other, better maintained ontologies. Like the resources that it supersedes, CONSO is not meant to be a high confidence resource, but rather an intermediate stage for curation as we find it a more appropriate home.
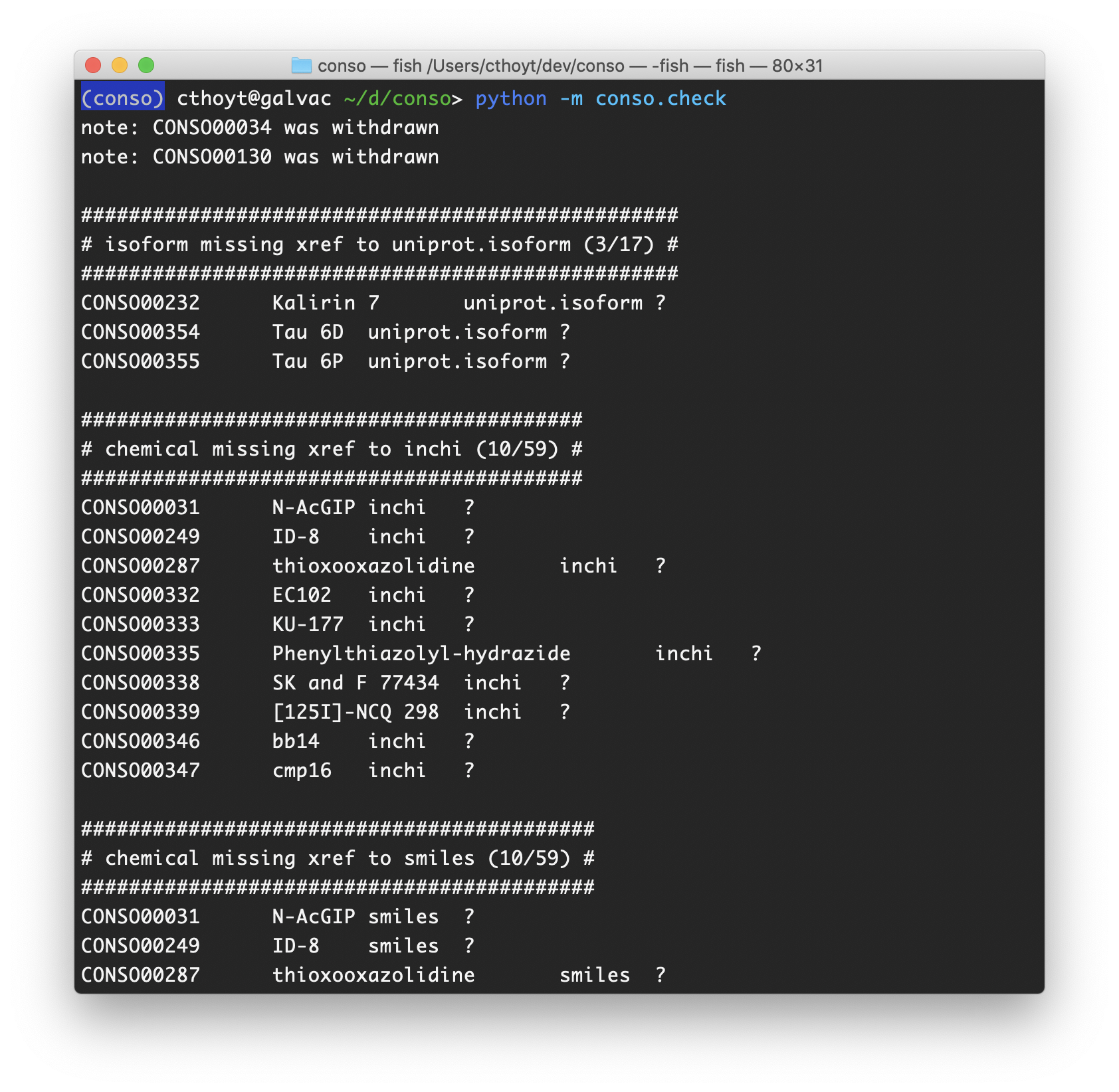
CONSO uses continuous integration on Travis-CI and an extensible verification script to check the integrity of the ontology on each push. The example above shows that isoforms are checked to have a relationship to their reference protein and that chemicals should be annotated with InChI, SMILES, and also InChI-key (not shown). There are also checks that all chemicals have been assigned chemical roles from the ChEBI ontology or CONSO (we’ve curated a few ourselves that get put in ChEBI later), that antibodies have been annotated with their targets.
It also performs simple checks that new terms are added with the next consecutive identifier, that minimum appropriate metadata has been added for new terms (we set the bar very high), and that all sheets use valid CONSO identifiers. Overall, these kinds of checks make it much easier to work in a team. As CONSO was curated by eight authors, they lessened the burden on explaining the rules for curation to new contributors - all changes must be made as pull requests and they must pass the tests on Travis-CI. If they don’t, new contributors just need to follow the instructions given to make updates until they do.
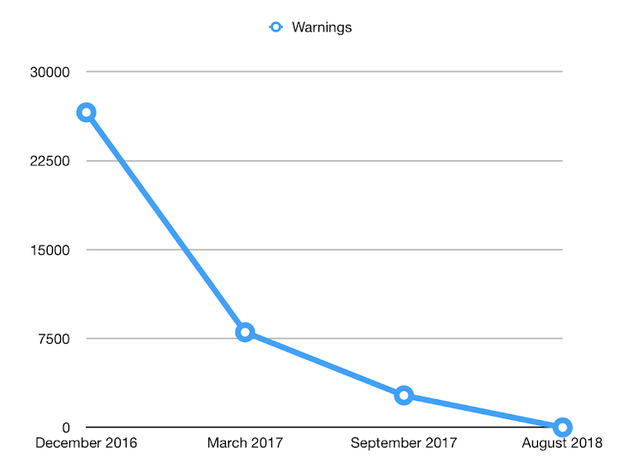
As the original goal of CONSO was to support the re-curation and enrichment of
NeuroMMSig, it’s nice to see in the above figure how its development allowed us
to reduce the number of BEL warnings in NeuroMMSig from almost 30,000 to zero
(in addition to some other BEL-specific fixes). This figure comes from and old
blog post on the Fraunhofer Human Brain Pharmacome site (which appears to have
been taken down?) and
my Ph.D. defense -
and will hopefully make it to primetime in the next release of NeuroMMSig
itself. Further, we were able to improve the overall quality of NeuroMMSig by
normalizing many terms that were redundant across namespaces as previously
described. CONSO was created using the guide in my previous blog
post. More specifically, the
experiences with CONSO inspired that guide to highlight the things that worked
well (and not the things that didn’t). In the end, CONSO is automatically
converted into an
OWL file, an
OBO file, a
BEL namespace
for BEL curation, and a website for
search and exploration that is automatically deployed with GitHub Pages. It’s
archived in Zenodo as well at
.
CONSO could not have been done without all of the hard work of the curators at Fraunhofer during the AETIONOMY project that provided the basis for this work. However, I would like to make a special thanks to my curation team - Rana Aldisi, Lingling Xu, Sandra Spalek, Esther Wollert, Kristian Kolpeja, and Yojana Gadiya for their immense contributions, Daniel Domingo-Fernández for helping when I needed it, and Stephan Gebel for learning so much so quickly in order to take over the curation program after I finished my Ph.D. and left Fraunhofer.