Adding New Literature Sources to the Wikidata Integrator
Scholia is a powerful frontend for summarizing authors, publications, institutions, topics, etc. that draws content from Wikidata. However, the content that’s available in Wikidata depends on what has been manually curated by community members and what has been (semi-) automatically imported by scripts and bots. The Wikidata Integrator from the Su Lab at Scripps automates the import of bibliometric information from Crossref and Europe PMC. This blog post is about how I added functionality to it to import from three prominent preprint servers in the natural sciences (arXiv, bioRxiv, and ChemRxiv) that can serve as a guide to others who want to have content about their field included with this tool.
A while back, I wanted to put an arXiv paper about the role of metadata in reproducible computational research that I co-authored with Jeremy Leipzig (@jermdemo) on Wikidata so it would appear on my Scholia page, but I didn’t want to do it by hand. I had already had good experiences using Magnus Manske’s SourceMD, Author Disambiguator, and related tools for queueing import of peer-reviewed papers with PubMed identifiers or digital object identifiers (DOIs), ORCID identifiers of co-authors, and ultimately assisted disambiguation of author names.
The only problem was that the SourceMD tool doesn’t deal with preprints. As an aside, both bioRxiv and ChemRxiv assign DOIs to their preprints that can be queried through Crossref, but this doesn’t exactly solve the problem of handling preprints in a special way, and it doesn’t solve the problem for arXiv. I asked around on Twitter and was turned towards the Wikidata Integrator project, which already supported building both general pipelines for automating content import in Wikidata and a specific one for publications. All I had to do was figure out how it works, and get hacking! I ended up sending the following three pull requests:
| Server | Pull Request |
|---|---|
| arXiv | SuLab/WikidataIntegrator#140 |
| bioRxiv | SuLab/WikidataIntegrator#169 |
| ChemRxiv | SuLab/WikidataIntegrator#140 |
At the time of writing, the arXiv and bioRxiv ones have been accepted, and the
ChemRxiv one is waiting for review - I think the maintainer was busy at a
conference this week, and now I’m writing on a Saturday. In another unrelated
pull request to the project, I added a vanity CLI, so when you install the code
with pip install wikidataintegrator, a program
wikidataintegrator-publication is installed for direct usage from the shell.
It can be used like this:
$ wikidataintegrator-publication --idtype arxiv 2101.05136
$ wikidataintegrator-publication --idtype biorxiv 2020.08.20.259226
$ wikidataintegrator-publication --idtype chemrxiv 13607438
This resulted in the following three Wikidata pages:
- arXiv: Q104846171
- bioRxiv: Q104920313
- ChemRxiv: Q104931192
If you’re using a DOI or PubMed identifier as the --idtype, you also have to
specify a --source, but since each of arXiv, bioRxiv, and ChemRxiv have their
own custom sources, this isn’t necessary. The program will print the Wikidata
identifier (starting with a Q followed by some numbers) of the newly created
item, or an error message if there was a problem. It’s quite smart and avoids
creating duplicate pages by checking if the ID type has already been used with a
pre-defined Wikidata property that goes with it. More on that below in the
tutorial, since we have to make that definition when adding new sources.
Implementing a New Importer
Luckily, all the work in implementing a new importer happens in one python
module:
wikidataintegrator.wdi_helpers.publication
. You can begin by clicking the
edit
button, which will automatically fork the repository and create a new branch.
I’m terrible using git and managing multiple remotes, so this is my preferred
way to start a PR in any repository that I might want to PR more than once.
Adding the right metadata

The first step is to identify the Wikidata property corresponding to entries in
your new source. This was already the case for arXiv
(P818) and bioRxiv
(P3951), so I added an entry to
the ID_TYPES dictionary in the Publication class where the key is the name
of the source (not necessarily the name on the property, keep it short and
simple).
The second step is to identify the Wikidata item corresponding to the source
itself and add it to the SOURCES dictionary in the Publication class (just
below the ID_TYPES dictionary).
Implementing the getter
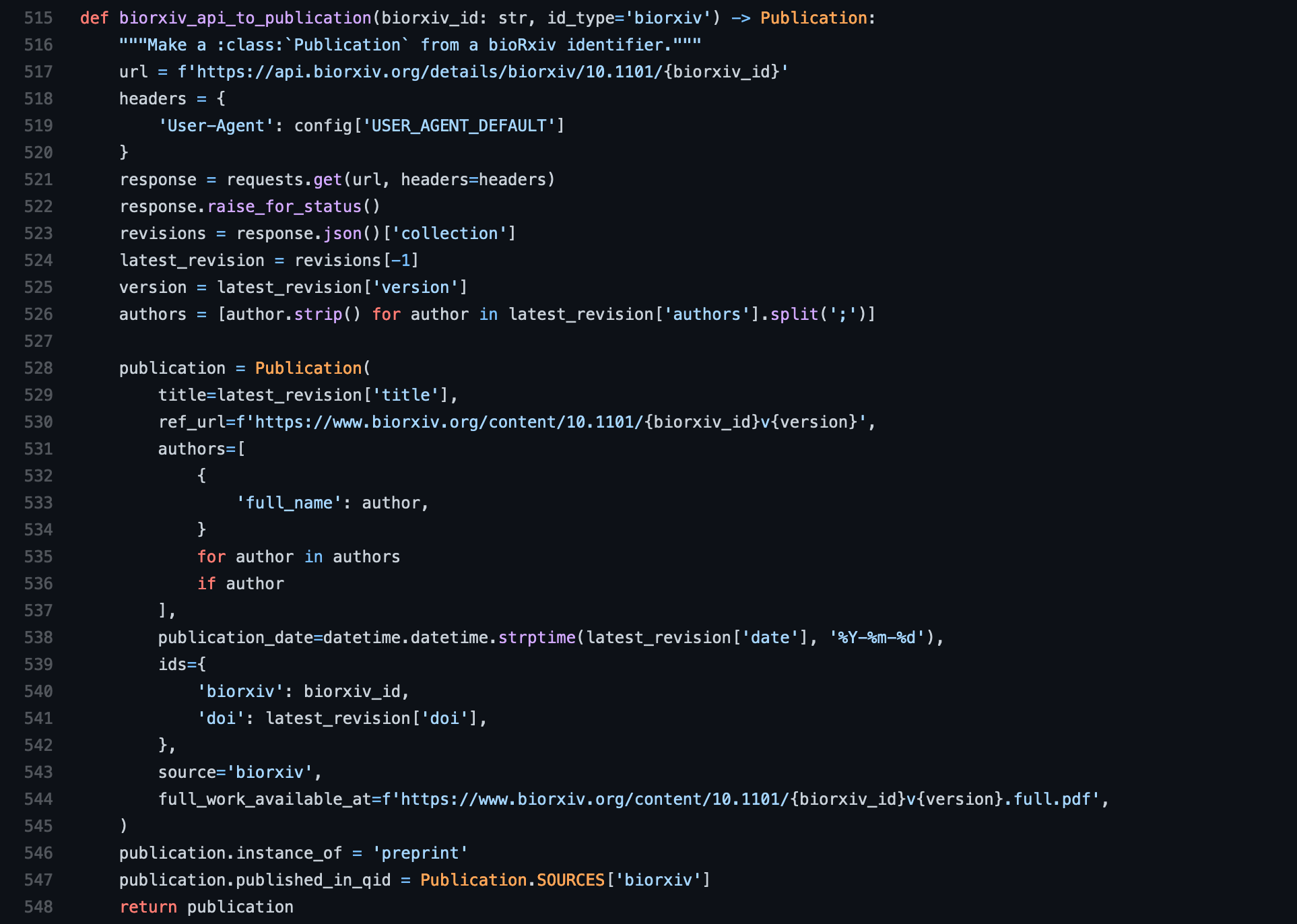
The third step is where the domain logic about your source comes in. You need to
implement a function that takes in an identifier that spits back an instance of
the Publication class. It also has to have an id_type argument where the
default value matches to the key you used in the sources. This actually isn’t
used anywhere, but must be there because of the interface that consumes it. This
function can live towards the bottom of the Python file and isn’t inside a
class. It’s best to put it next to the bioRxiv, arXiv, and Crossref ones.
Most of the way you get data from your source is up to you. Most sources have some kind of endpoint that can be queried and returns JSON - note that Wikidata Integrator has a consistent USER AGENT that tells services what kind of code is querying it. This is important if you’re hitting an API with many queries so sysadmins can see what’s going on.
The Publication class is pretty self-explanatory except for a few parts.
- The
authorskeyword arguments takes a list of dictionaries whose keys arefull_nameandorcid. You can omitorcidor passNone. bioRxiv isn’t currently providing author information like ORCID identifiers in its API, so it did not show up in this example, but you can see in the other parts of the code for PMC and PubMed how this works. - Make sure that the keys in the
idskeyword argument correspond to keys inID_TYPES. If your document has more than one ID, you can put them her (though there is some debate whether this is a good idea). - You have to set the
instance_ofproperty not with the__init__()because it gets handled with a Python descriptor. This is a design choice in the Publication class that is out of our hands. - Don’t forget to set the
published_in_qidwhich will correspond to thevenueon your wikidata item.
Tying it all together
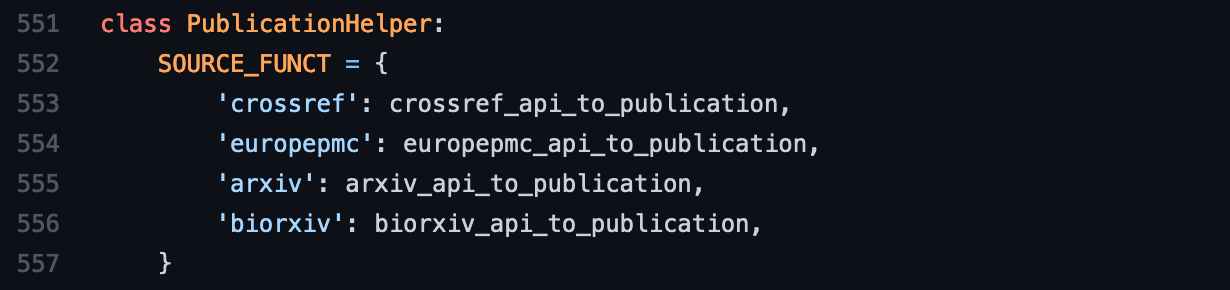
In the fourth step, you need to register your newly implemented function in the
SOURCE_FUNCT dictionary inside the PublicationHelper class. Make sure the
key you use is consistent with before, and use the function as the value. Don’t
use parentheses, since you want to actually have the function be the value and
not the result of calling the function. This is part of a programming paradigm
called “functional programming”.
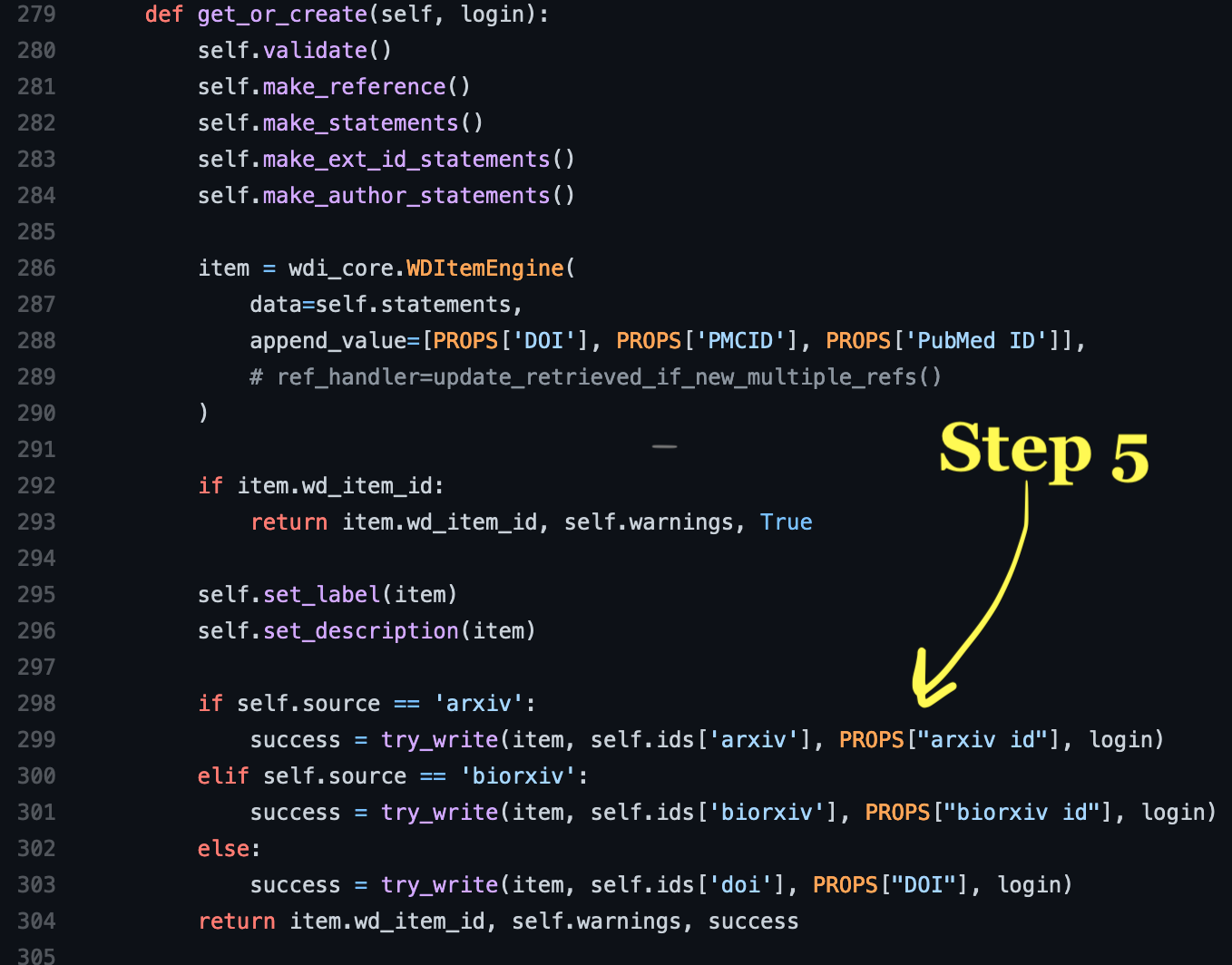
In the fifth and final step, you need to scroll up to the get_or_create()
function in the Publication class to tell it which ID is the primary key for
your item. Add a new conditional elif self.source == 'your key' to check for
your source key (same as all the other places you added it) then add the key
corresponding to your source from the ids (this was the part from step 3) and
the correct property. The previous examples use PROPS which is redundant. You
can use self.ID_TYPES['your key'].
Testing
Try running the following code with a valid ID and hope that everything works! If you are having issues here, then you can always send a draft pull request to solicit help from the maintainers of the project.
$ wikidataintegrator-publication --idtype "your key" "your id"
Be careful here, since this will hit the live Wikidata instance. If you make a new item that has a problem, please try to fix it since deleting entries from Wikidata isn’t so common, and we don’t want to add to the mess!
My Source Doesn’t Have a Wikidata Property
In the case of ChemRxiv, DOIs are available for each article, so I did not need
to add a new entry to ID_TYPES dictionary. However, scholarly articles on
Wikidata typically use the DOI to point to the peer-reviewed article, and a
preprint-specific property to point to the preprint describing the same paper (I
know, confusing…).
I created a property proposal for “ChemRxiv ID” on Wikidata to help rectify this. You can propose a new property from this page but beware: Wikidata property maintainers are quite cautious to add new things and aren’t necessarily giving the most prompt feedback.
I had a lot of fun working on this new codebase and this blog post, and it was a reminder of the nice discussion I had with Andrew Su last year. I hope this post enables others to add support for medRxiv, Preprints.org, and other places where people are leaving their pre-prints!