How to Curate the INDRA Database
With the recent paper on Gilda and approaching INDRA 2 and INDRA database papers coming up, I’ve put together a visual guide on how to curate statements extracted by INDRA through the web interface at https://db.indra.bio.
Navigate to the INDRA database portal using this link.
Register for an account if you don’t already have one by clicking the register box.
Login with your email/password.
Enter search text. This can be a gene symbol, chemical, or any other biomedical entity. In this tutorial, we’ll search for “AKT1.”
Now, click the “Ground with Gilda” button. Gilda will automatically look up the most likely database identifier that goes with your search string. It’s quite smart and can even disambiguate different senses of the same word. If it’s not sure, it will ask you to pick from a list.
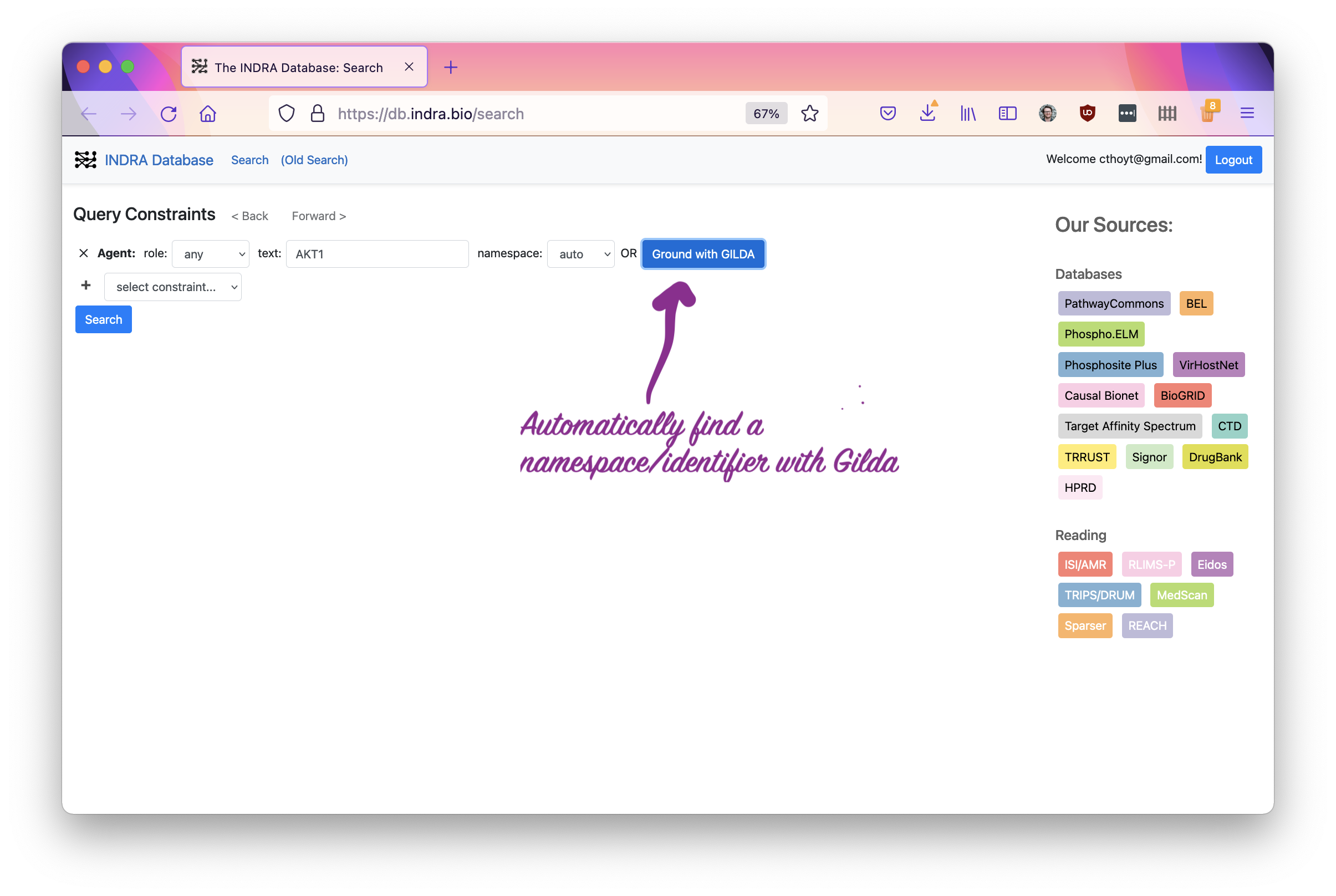
In this example, I chose AKT1, which was pretty easy for Gilda to ground and didn’t need me to check. It also reports the confidence in the grounding and the namespace to which it grounded in the box. Click the search button to get going to the next step!
Now you will see the search results. On the left, it has many kinds of statements that INDRA models. They’re hierarchical, meaning you can click on one to expand to more specific statement types.
On the right are badges for the different sources that give evidence for each statement. The ones with the black text correspond to databases, like BioGRID, and the ones with white text correspond to reading systems, like REACH.
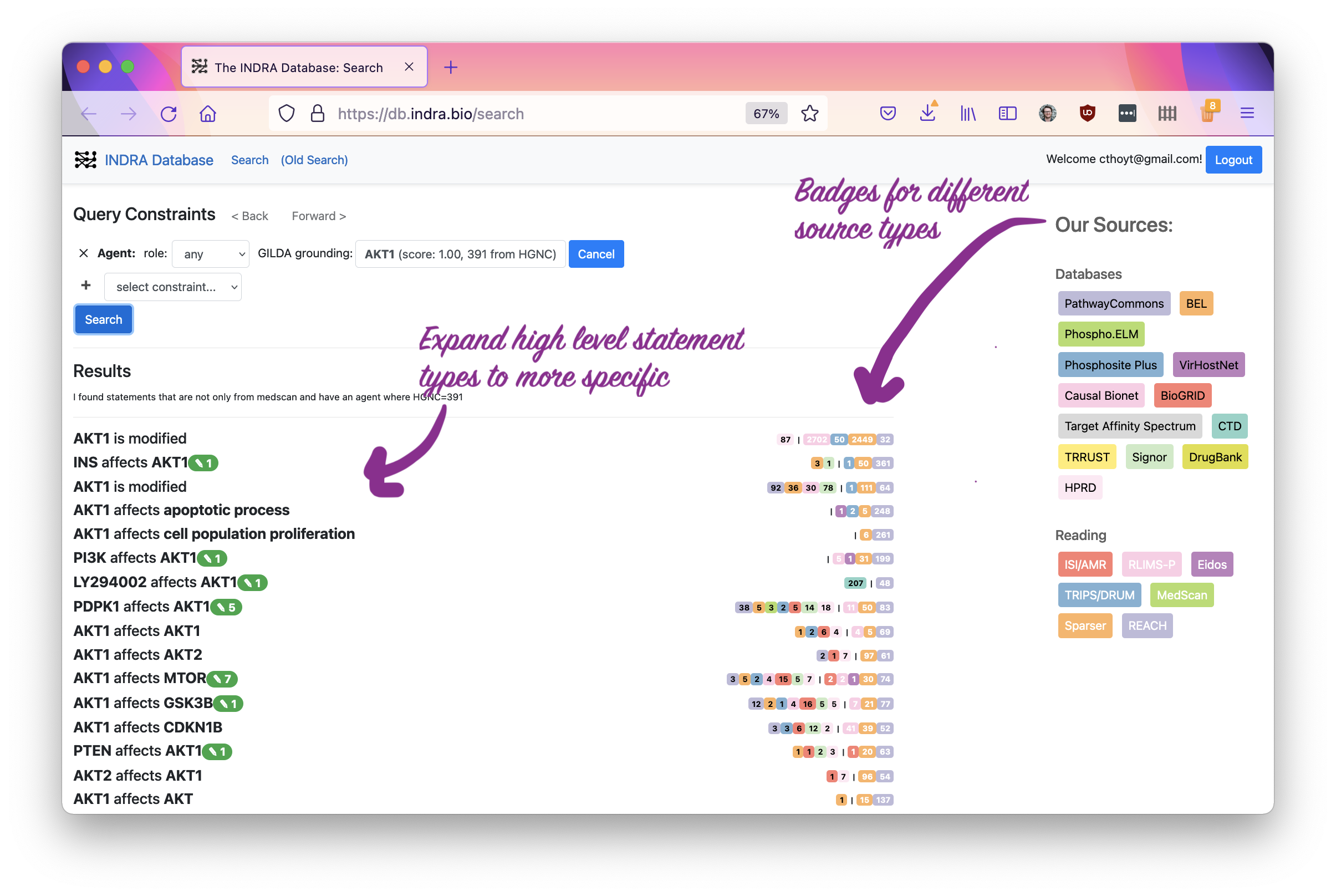
The next image shows the expansion of AKT affects BAD to
AKT1 phosphorylates BAD. Note that the third level is the same as the second -
this is because there are actually some more specific phosphorylation events
contained in here as well!
Open up the curation interface by pressing the pencil button next to the evidence you want to curate. Note that some sources, like BioPAX (from Pathway Commons) don’t give evidence text, so these aren’t appropriate for curation via the INDRA Database.
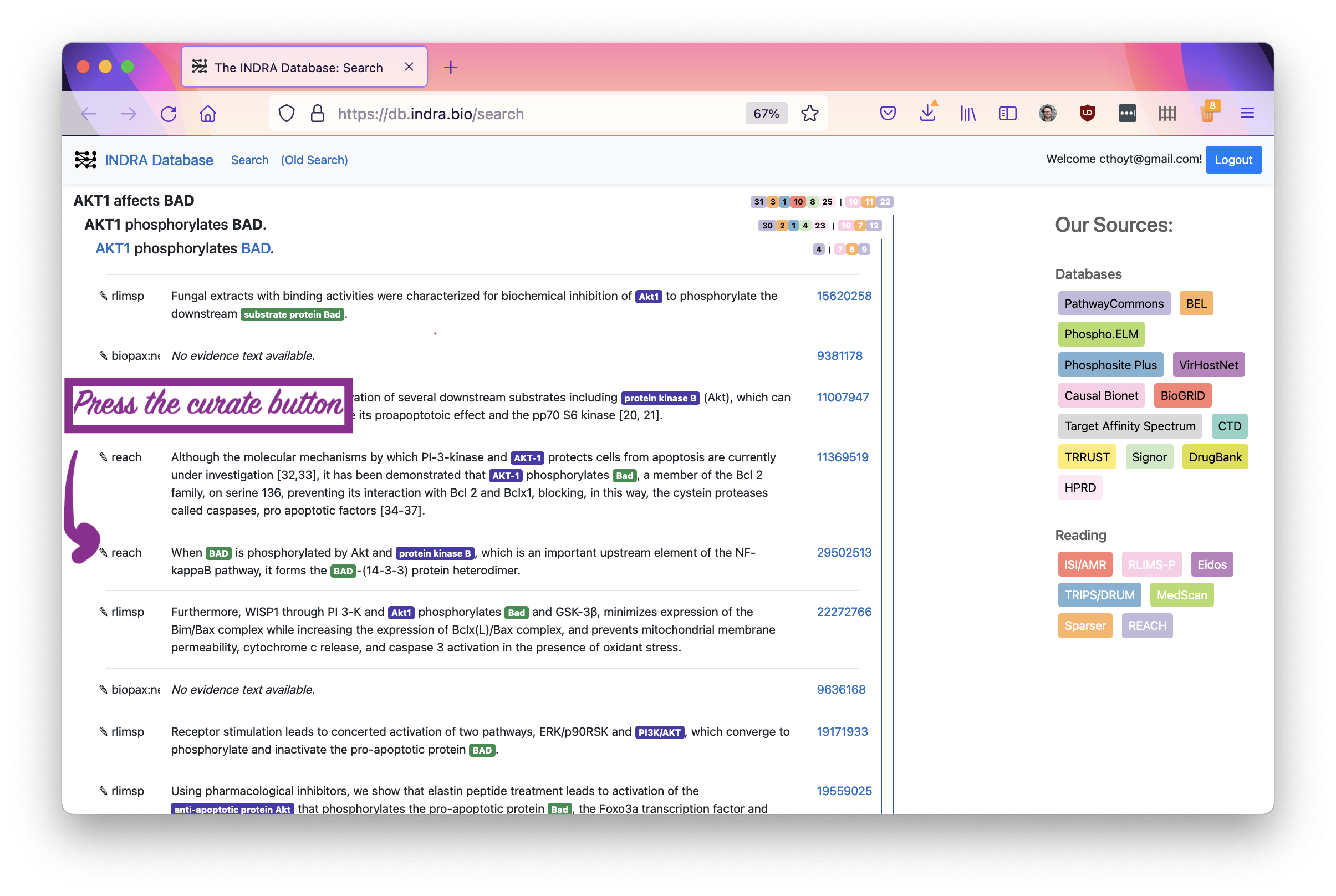
Now that the curation menu has come up, you can select one of several error types. Use your best judgement if a statement is really correct. Note that INDRA does synonym disambiguation, so the label for the statement may not match to the highlighted text. You can click the entity names in the statement header to open pages with more information about the entities, including their synonyms.
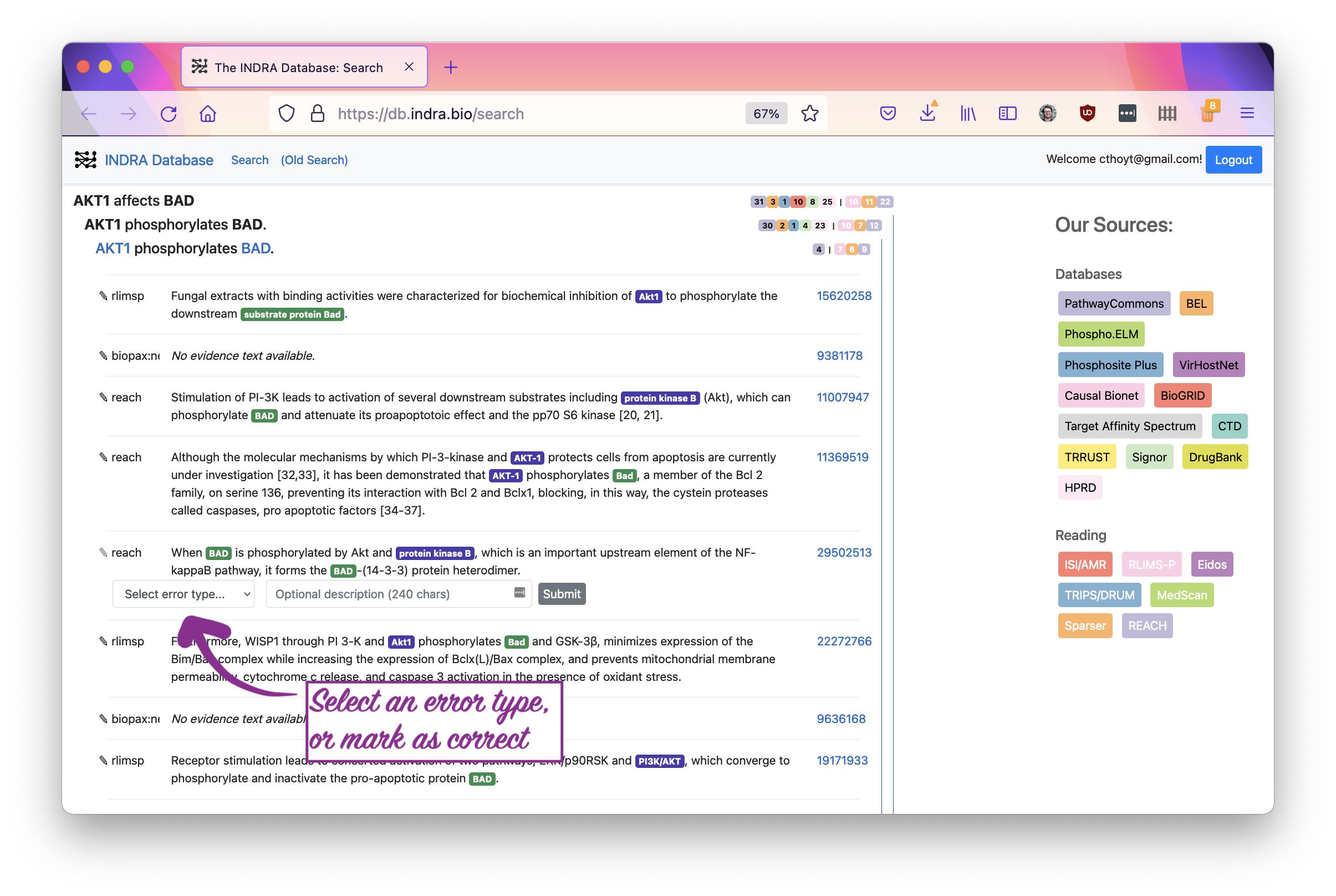
If you want to leave a note that explains why you made the curation you did, that would be very helpful! Finally, smash that submit button.
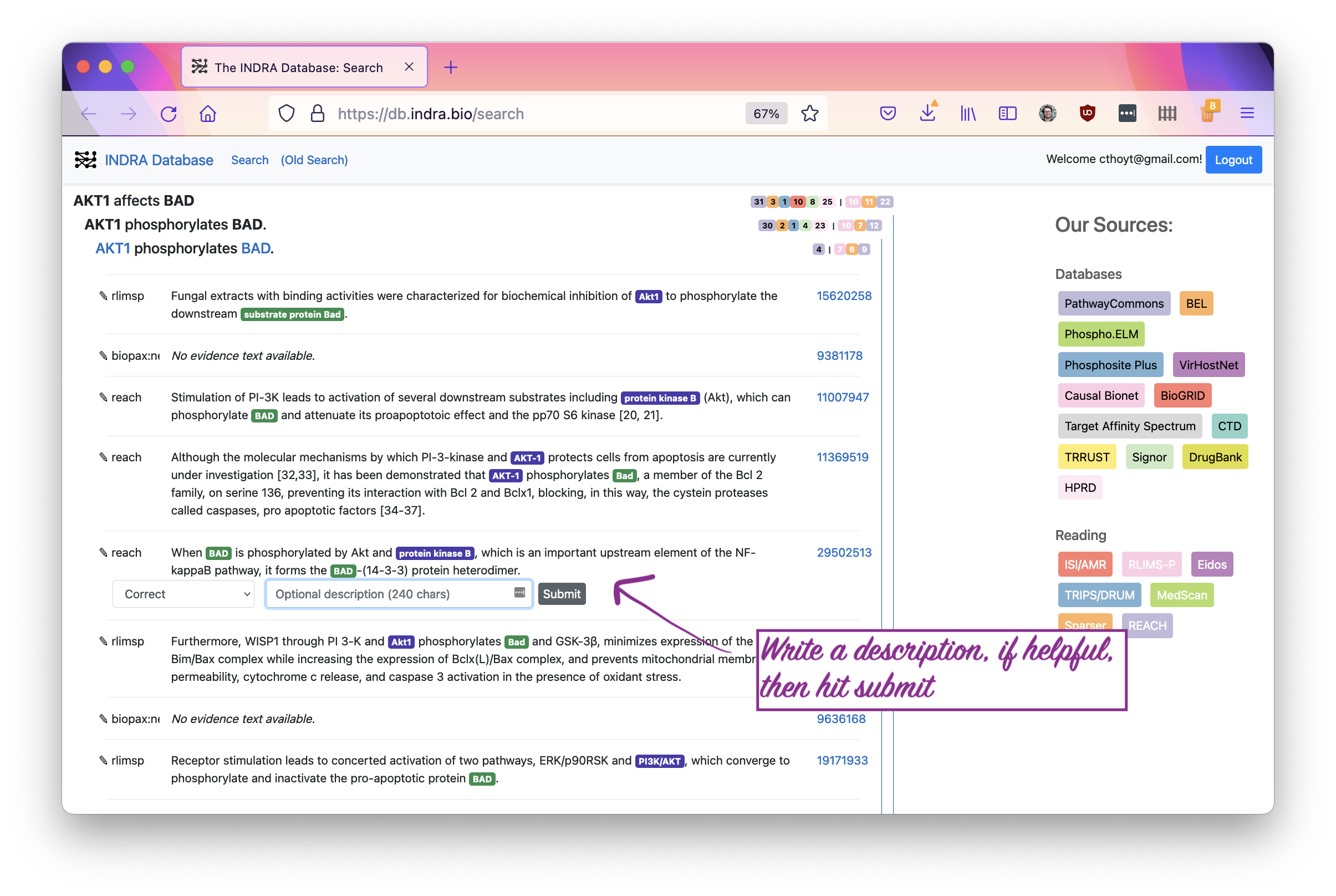
You’ve now contributed a curation! Thank you very much. You and the rest of the scientific community will now disproportionately benefit from this small amount of effort because of the large-scale extraction efforts of the INDRA Database and combination with other curations.
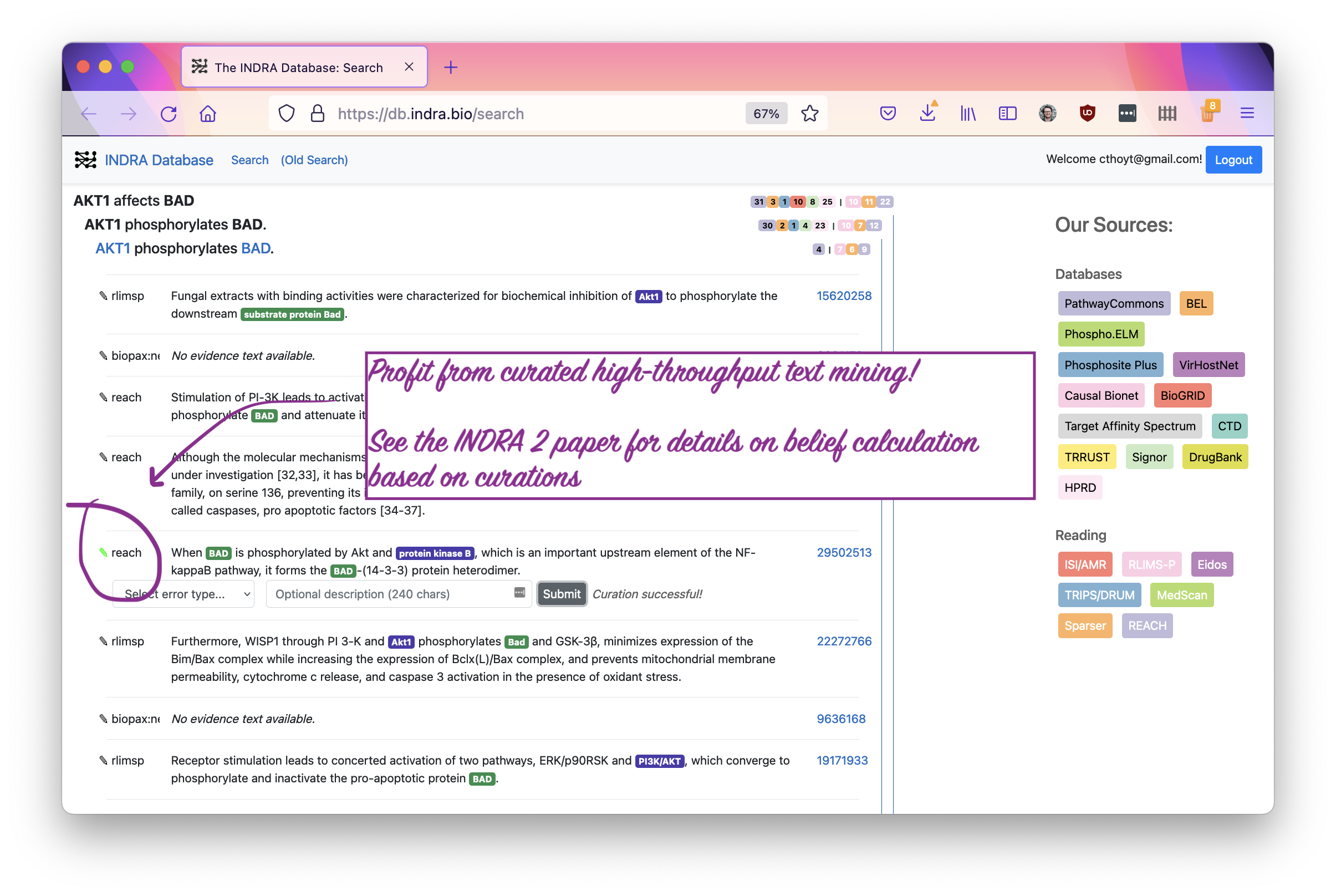
Stay tuned for the upcoming INDRA 2 paper that outlines how curations can be
used to assess the quality of each statement (at each level of hierarchical
abstraction, too) as well as the INDRA Database paper describing this resource.
We’re currently working on
updating the API for bulk
downloading curations from the INDRA Database and for use with the INDRA
assemble corpus utility
indra.tools.assemble_corpus.filter_by_curation().
Update Here’s the INDRA 2 preprint:doi:10.1101/2022.08.30.505688