Global Core Biodata Resources in the Bioregistry
The Global Biodata Coalition released a list of Global Core Biodata Resources (GCBRs) in December 2022, comprising 37 life science databases that they considered as having significant importance (selected following this procedure). While the Bioregistry does not generally cover databases, many notable databases have one or more associated semantic spaces that are relevant for inclusion. Accordingly, 33 of 37 of the GCBRs (that’s 89%) have one or more directly-related prefixes in the Bioregistry. This post gives some insight into this landscape.
Background on the Bioregistry
The Bioregistry is a catalog of identifier schema for concepts in the life and
natural sciences. These identifier schemata often arise from databases that
create stable, locally unique identifiers for a given entity type. For example,
the Universal Protein Resource (UniProt) creates stable, locally unique
identifiers for proteins such as P0DP23
for Calmodulin-1. Similarly, the Chemical Entities of Biological Interest
(ChEBI) creates stable, locally unique identifiers for chemicals such as
138488 for alsterpaullone. The
Bioregistry contains records about these identifier schema including the regular
expression pattern that can be used to validate locally unique identifiers
(e.g., the UniProt one is quite complicated but the ChEBI one is simply a string
that looks like a number ^\\d+$), the prefix that should be used when
constructing compact URIs (CURIEs) (e.g., uniprot for UniProt), a URI format
string that can be used to convert the local unique identifier into a URI (e.g.,
for usage in semantic web applications), and other useful metadata for the
standardization of the identification of life and natural sciences concepts.
Some databases induce more than one identifier schema. For example, in addition
to the identifier schema for proteins, UniProt also has disjoint identifier
schemata for
subcellular locations,
diseases, and several others.
In the case of UniProt, the main identifier schema is for proteins, and is
therefore given the same prefix as the name of the database (i.e.,
uniprot). The prefixes for additional
identifier schemata are constructed as subspaces using a dot-delimiter.
Alternatively, some databases that mint multiple identifiers schemata, such as
the
Clinical Interpretation of Variants in Cancer (CIViC)
database, do not have a “main” entity type and therefore use subspaces for all
of its prefixes (e.g., civic.gid for
genes, civic.vid for variants, etc.)
Back to GCBRs
It’s important to note that the Bioregistry maintains records for the identifier schemata, and not the databases themselves. Other catalogs like Wikidata and FAIRsharing already do an excellent job of maintaining records on databases and other larger efforts. With this background out of the way, we can return to the main question of this post: how do the Global Core Biodata Resources (GCBRs) relate to the Bioregistry? I stratified the list of 37 into four categories:
- 19 databases that have a single identifier schema (i.e., correspond 1-to-1 with Bioregistry records)
- 12 databases that have multiple identifier schemata (i.e., correspond 1-to-many with Bioregistry records)
- 2 databases that have a more complicated relationship to Bioregistry records
- 4 databases that don’t have identifier schemata (this turns out to be a very short list!)
GCBRs with a Single Identifier Schema
The following 19 databases in the GCBR list have a one-to-one correspondence with a Bioregistry prefix. In some cases, this categorization is partly subjective as many of these databases’ curators are heavily involved in other related efforts that their databases heavily reuse. For example, the Zebrafish Information Network is a model organism database that is heavily involved in the curation of ontologies for zebrafish anatomy and development (see zfa), developmental stages (see zfs), and phenotypes (see zfa).
| Database | Bioregisry Prefix |
|---|---|
| Alliance of Genome Resources Knowledge Base | agrkb |
| Bacterial Diversity Metadatabase | bacdive |
| Chemical Entities of Biological Interest | chebi |
| EcoCyc | ecocyc |
| VEuPathDB ontology | eupath |
| Global Biodiversity Information Facility | gbif |
| Genome Aggregation Database | gnomad |
| Gene Ontology | go |
| InterPro | interpro |
| Mouse Genome Informatics | mgi |
| PDB Structure | pdb |
| European PubMed Central | pmc |
| PomBase | pombase |
| ProteomeXchange | px |
| Reactome | reactome |
| Rhea, the Annotated Reactions Database | rhea |
| Saccharomyces Genome Database | sgd |
| UCSC Genome Browser | ucsc |
| Zebrafish Information Network Gene | zfin |
GCBRs with Multiple Identifier Schemata
The following 12 databases in the GCBR list have identifier schemata and therefore correspond to multiple Bioregistry records. Note, this list might be incomplete in cases where there are other relevant identifier schemata that haven’t been added to the Bioregistry. If you’re aware of one, please let me know or send a new prefix request!
| Database | BioregistryPrefix | Name |
|---|---|---|
| CIViC | civic.aid |
CIViC Assertion |
civic.did |
CIViC Disease | |
civic.eid |
CIViC Evidence | |
civic.gid |
CIViC Gene | |
civic.sid |
CIViC Source | |
civic.tid |
CIViC Therapy | |
civic.vid |
CIViC Variant | |
| BRENDA | brenda |
BRENDA Enzmye (duplicate of eccode) |
brenda.ligand |
BRENDA Ligand | |
brenda.ligandgroup |
BRENDA Ligand Group | |
bto |
BRENDA Tissue Ontology | |
| ChEMBL | chembl |
ChEMBL |
chembl.compound |
ChEMBL Compound (subspace) | |
chembl.target |
ChEMBL Target (subspace) | |
| Ensembl | ensembl |
Ensembl Gene, Transcript, etc. |
ensemblglossary |
Ensembl Glossary | |
| FlyBase | flybase |
FlyBase Gene |
fbbt |
Drosophila Gross Anatomy | |
fbcv |
FlyBase Controlled Vocabulary | |
fbrf |
FlyBase Reference Report | |
fbsp |
Fly Taxonomy | |
fbtc |
FlyBase Cell Line | |
| HGNC | hgnc |
HGNC Gene |
hgnc |
HGNC Gene Group | |
| PANTHER | panther.family |
PANTHER Family |
panther.node |
PANTHER Node | |
panther.pathway |
PANTHER Pathway | |
panther.pthcmp |
PANTHER Pathway Comparison | |
| PharmGKB | pharmgkb.disease |
PharmGKB Disease |
pharmgkb.drug |
PharmGKB Drug | |
pharmgkb.gene |
PharmGKB Gene | |
pharmgkb.pathways |
PharmGKB Pathway | |
| Orphanet | orphanet |
Orphanet |
orphanet.ordo |
Orphanet Rare Disease Ontology | |
| Rat Genome Database | rgd |
Rat Gene |
rgd.qtl |
Rat Quantitative Trait Loci | |
rgd.strain |
Rat Strain | |
| UniProt | uniprot |
UniProt Protein |
uniprot.arba |
UniProt Association-Rule-Based Annotator | |
uniprot.chain |
UniProt Chain | |
uniprot.disease |
UniProt Disease | |
uniprot.isoform |
UniProt Isoform | |
uniprot.keyword |
UniProt Keyword | |
uniprot.location |
UniProt Location | |
uniprot.proteome |
UniProt Proteome | |
uniprot.ptm |
UniProt Post-translational Modification | |
uniprot.resource |
UniProt Resource | |
uniprot.tissue |
UniProt Tissue | |
uniprot.var |
UniProt Variant | |
uniparc |
UniProt Archive | |
uniref |
UniProt Reference Clusters | |
| Wormbase | wormbase |
Wormbase Gene |
wbrnai |
Wormbase RNAi |
GCBRs with a Confusing Relationship to Identifier Schemata
This list only gets two databases:
The reason that the DDBJ and ENA (and also NCBI GenBank, but it was not included as a GCBR) have a complicated relationship is because of their involvement in the International Nucleotide Sequence Database Collaboration (INSDC). This is a coordination effort between the DDBJ, EMBL-EBI and NCBI to promote interoperability between nucleotide sequence and related databases. Here’s a screenshot from their website that explains how certain services interact across these platforms:
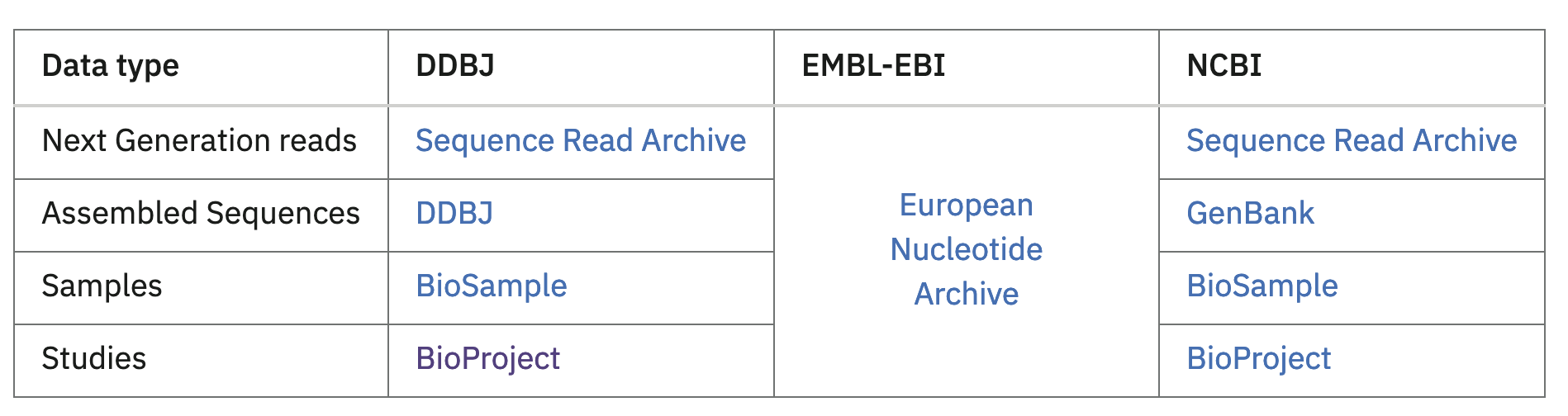
Modeling this in the Bioregistry has proven very tricky. There are several ongoing discussions related to clarifying the relationships between these databases and various identifier schemata in the Bioregistry’s issue tracker (see #108, #118, and #131). Here are a few of the relevant prefixes:
ena.embl(ENA-specific)bioproject(DDBJ-specific)genbank(NCBI-specific)insdc.run(non-specific)
If you’ve got some insight into these resources, please join for discussion on the Bioregistry issue tracker.
GCBRs with No Identifier Schemata
The four GCBRs that contain no identifier schemata have a common attribute: they all reuse other identifier schemata.
- Clinical Genome Resource (uses
hgnc,mondo,pharmgkb.pathways, etc.) - GENCODE (uses
genbank) - GWAS Catalog (uses
dbsnp,hgnc.symbol, andefo) - STRING (uses
uniprot)
Interestingly, since these resources transitively use GenBank, dbSNP, and the Experimental Factor Ontology but these databases were not themselves included in the list of GCBRs. There are several other examples of databases transitively used by other GCBRs appearing in other parts of this categorization where this is also true. Without exhaustively going through all four resources, I thought I’d use the GWAS Catalog to illustrate how it looks when a database re-uses other identifier schemata-providing databases.
First, a genome-wide association study (GWAS) identifies statistical correlation between genomic markers such as single nucleotide polymorphisms (SNPs) and disease, phenotypes, or other traits across a large and diverse population, usually on the scale of thousands to tens of thousands of individuals. The GWAS Catalog is an EMBL-EBI database of published GWASs, their metadata (e.g., experimental design), and the statistically significant associations they identified. It maintains information about the following kinds of things:
| Type | Example | Vocabulary |
|---|---|---|
| Variant | rs7329174 | dbSNP (dbsnp) |
| Gene | ELF1 | HGNC Gene Symbols (hgnc.symbol) |
| Region | 2q37.1 | - |
| Trait | breast carcinoma | Experimental Factor Ontology (efo) |
A given association comprises a p-value for the association between a variant and a trait. Typically, there is a gene annotated to the SNP to make interpretation more simple, though this is no easy task. I’d suggest following Eric Faumann on Twitter (@Eric_Fauman) (or Mastodon if/when he moves there, because I’m not a big Twitter fan anymore) for interesting examples of this.
Interestingly, through the process of writing this post, I realized GWAS Catalog assigns such as GCST000858 which have their own unique semantic space and provider worthy of an entry in the Bioregistry, but that will probably have to wait until after winter break.
Afterthoughts
I’ve got a few parting comments on the construction of the list of GCBRs:
- There appears to be an over-representation of gene/genome and model organism databases. This probably reflects the fact that these are the oldest kinds of bioinformatics resources. Overall, the list does include a nice variety of different kinds of resources.
- There appears to be an over-representation of databases from the EBI, SIB, and NCBI. Since some of the criteria for inclusion were to demonstrate existence for more than five years, this makes it difficult for most databases to even be considered, as most don’t have appropriate funding or governance models for longevity. It’s also likely that there’s significant overlap between the professional networks of members of these large, prominent institutions and the Global Biodata Coalition, which could have lead to increased interest and familiarity with certain resources that ultimately were accepted.
- There appears to be an over-representation of American and European resources. This might be partially due to the criteria that resources were required to have an English language version, but I think more generally it’s due to the unfortunate reality that there aren’t many reliable resources being developed and maintained in other locale for topics in scope for the GCBR list. I don’t think this is a reflection on a lack of interest for diversity and inclusion on the part of the Global Biodata Coalition. Further, efforts like INSDC seem like a good idea to coordinate efforts outside of the typical western research bubble, and the inclusion of the DDBJ in the list seems to pay deference to this sentiment.
While these observations might be thought-provoking, I’ve only made them at a subjective level. I think a more interesting follow-up would to be to use linked data approaches (e.g., through Wikidata) to aggregate relevant information together about each of these resources and make more objective summaries. To support that, I’ve created a collection on the Bioregistry bioregistry.collection:0000010 that contains the list of prefixes mentioned here.
Serendipitously, the scientific program committee of the 16th Annual International Biocuration Conference (Biocuration 2023) has invited the Executive Director of the Global Biodata Coalition, Guy Cochrane, to give a keynote presentation at the conference. This will be very timely, and I’m looking forwards to meeting him in person and chatting about this. If you’re a reader of my blog, you’re likely a creator, user, or both of resources like the one in the GCBR list. I’d suggest you consider submitting an abstract for the conference and meeting me in Padua in April!