Text-based embeddings of ontology terms
The Ontology Lookup Service (OLS) is now indexing
dense embeddings for ontology terms constructed from term labels, synonyms, and
descriptions using LLMs. I maintain a Python client library for the OLS
(ols-client) and was recently asked to
implement a wrapper to the OLS’s API endpoint that exposes these embeddings.
This post is a demo of how to use that code, and how I replicated the same
embedding functionality with PyOBO to
arbitrarily extend it to ontologies and databases not in OLS.
I’ve been working on modeling clinical trials in collaboration with Sebastian Duesing at the Ontology for Biomedical Investigations (OBI), so I’m going to use the OBI term for clinical trial (OBI:0003699) that we recently minted together as an example.
Embeddings from OLS
Below is a screenshot of the OLS page for this term, as of August 4th, 2025:
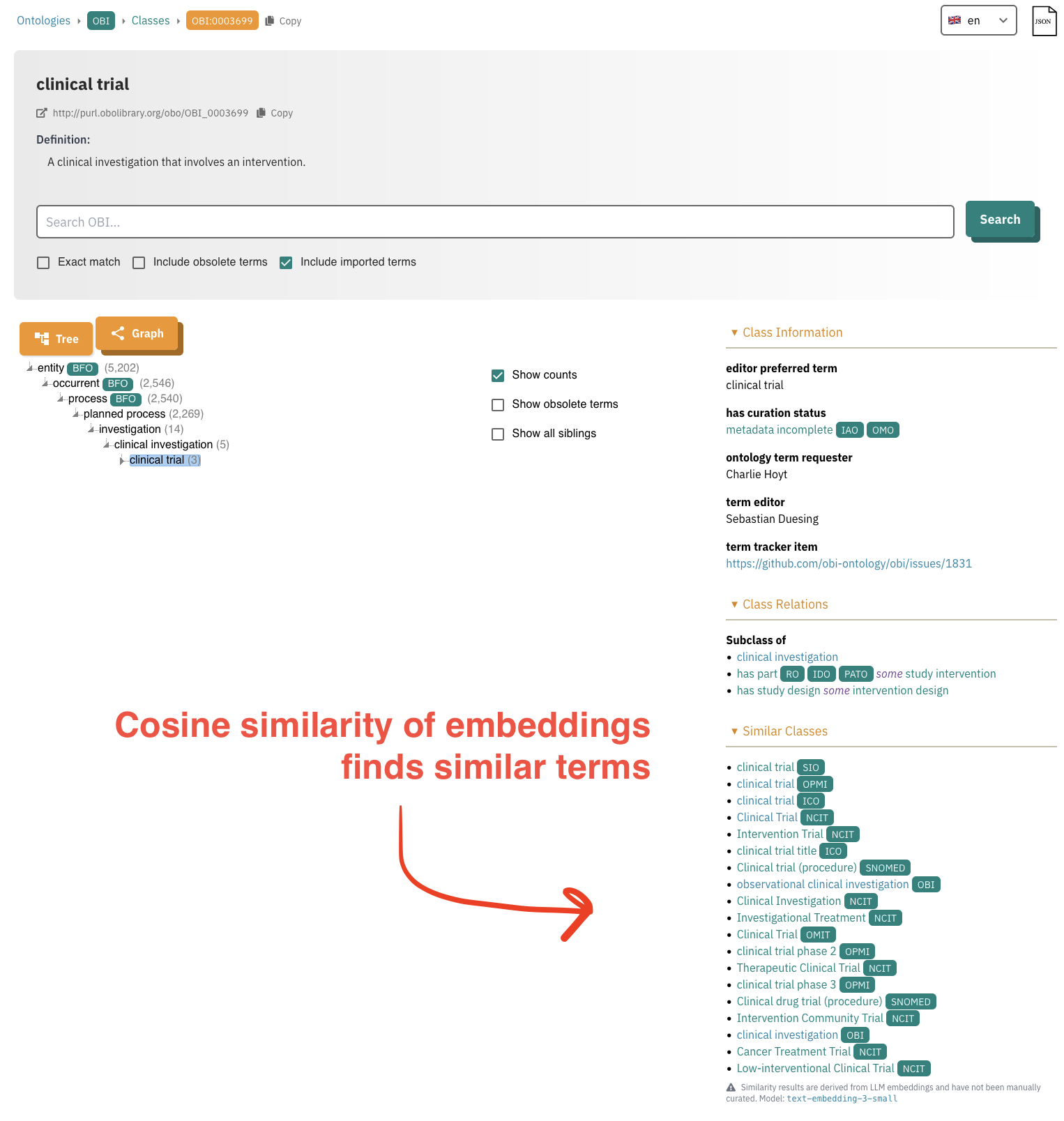
There’s a newly added section on similar terms. Here’s what it says about these:
Similarity results are derived from LLM embeddings and have not been manually curated. Model: text-embedding-3-small
From a first glance, the results here have very high precision. This shouldn’t be surprising, considering that labels, synonyms, and descriptions contain very high signal. Of course, there’s room for arguing about the nuance of ontological differences, but this is a largely unhelpful discussion in my experience when the goal is to do ontology merging and data integration. In my recent post where I described the landscape of clinical trial modeling in the OBO Foundry and related biomedical ontologies, I actually had already curated several of these semantic mappings by hand, which I’m hoping to add to OBI via SSSOM. Looking forward for the OLS, it would be great if there were a mini curation interface where these could be confirmed or rejected as exact mappings, and persist the resulting curations as SSSOM.
There was a request to expose the embeddings via my OLS client package, which I solved with only a few lines of code here. Now, you can get the embedding for clinical trial (or any other term, based on the ontology/IRI combination) as a list of floating point numbers:
from ols_client import EBIClient
client = EBIClient()
embedding: list[float] = client.get_embedding(
"obi", "http://purl.obolibrary.org/obo/OBI_0003699"
)
The next step is to be able to calculate the (cosine) similarity between two terms, which can be done between the OBI term for clinical trial, and the National Cancer Institute Thesaurus (NCIT) term for clinical trial as a single floating point number:
from ols_client import EBIClient
client = EBIClient()
similarity: float = client.get_embedding_similarity(
"obi",
"http://purl.obolibrary.org/obo/OBI_0003699",
"ncit",
"http://purl.obolibrary.org/obo/NCIT_C71104",
)
Embeddings from PyOBO
I think using an OpenAI model is a bit overkill for two reasons. First, there
are smaller, non-large language models like
BERT that can get
the same job done. Second, they’re free to download and can be run on commodity
hardware, versus needing to pay OpenAI for access to their embeddings.
Specifically, I’ve been looking at SBERT (Sentence-BERT),
which is a variant of the BERT architecture that works better on sentences and
can be easily used via the
sentence-transformers
Python package.
I’ve developed the pyobo package,
which gives unified access to ontologies and databases that are ontology-like.
It has functionality for getting the labels, synonyms, and descriptions for
terms in both.
It wasn’t difficult to re-implement the same functionality as the OLS in PyOBO such that it can be run locally on a larger variety of resources. Here’s the same lookup for text embedding and similarity:
import pyobo
>>> pyobo.get_text_embedding("OBI:0003699")
[-5.68335280e-02 7.96175096e-03 -3.36112119e-02 2.34440481e-03 ... ]
>>> pyobo.get_text_embedding_similarity("OBI:0003699", "NCIT:C71104")
0.24702128767967224
This could be improved with the ability to do batch lookup, which is probably the way people would want to use this functionality. Even better, because of how ML is implemented on GPUs and related hardware, batching effectively comes for free, only limited by memory contraints.
Text embeddings aren’t the end of the story - I’ve been working for several years on applications fo graph machine learning in biomedical applications. A lot of the good methodological and software engineering ideas I’ve had/encountered have gone into the PyKEEN Python software package. One of those ideas that jointly leverages text and knowledge graph embeddings is NodePiece, the work of PyKEEN core developer Michael Galkin. A guide on using it with PyKEEN is here, but there’s a lot more research to present on this than fits in the postscript of a blog post or a software tutorial. Along with the last few years of my work on data integration, I haven’t yet published my magnum opus on biomedical knowledge graph construction and applications with graph machine learning.