Measuring the impact of the Bioregistry
The Bioregistry is a database and toolchain for standardization of prefixes, CURIEs, and URIs that appear in linked (open) data. While I created it in 2019 as a component of PyOBO in order to support parsing database cross-references appearing in biomedical ontologies, it has since become an independent project with a community-driven governance model and much broader applications. This post is a first attempt to quantify its usage and impact.
What are Usage and Impact?
For a foundational resource like the Bioregistry, there are two kinds of usage. First, direct usage encompasses when a workflow directly reuses the Bioregistry’s data, software, or web application. For example:
- The SSSOM-py Python package for interacting with semantic mappings uses the Bioregistry for supplying a comprehensive default prefix map during parsing of SSSOM files.
- The BridgeDb identifier mapping service uses the Bioregistry Python package to test its source metadata are properly standardized (see here)
- The Protegé ontology editor uses the Bioregistry’s API to look up information about prefixes.
- WikiPathways uses the Bioregistry’s resolution service to linkify compact URIs ( CURIEs).
Second, indirect usage encompasses any other data, software, web application, etc. that builds on direct usages. For example, any ontology that is edited using Protegé indirectly uses the Bioregistry, like the Disease Ontology (DO).
I consider usage to be a very good proxy for impact, especially when considering indirect usage. Allen Baron et al. recently published The DO-KB Knowledgebase: a 20-year journey developing the disease open science ecosystem, which made a quantification of DO’s cumulative impact over the last decades. One area of this study focused on literature citations and resulted in the development of a reusable, open-source software package for such analyses. I don’t think anyone would disagree that the DO has had high impact.
I think that because DO is built using tools that rely on the Bioregistry, it’s fair to claim that DO’s (recent) impact is partially due to the Bioregistry.
The Bioregistry is in the excellent position where there are a variety of direct and indirect usages, many of which are highly impactful, meaning that the Bioregistry can share (a bit) in their glory. Later in this post, I’ll give a more quantitative justification for that statement.
Why Quantify Usage and Impact?
I think some of the main reasons for quantifying a project’s usage and impact are to:
- justify its continued maintenance and improvement
- get credit for making something important
- use previous usage as examples for increased adoption
- get funding to continue maintaining and improving it
At the moment, I am preparing to apply for the Bioregistry to be recognized by the Global Biodata Coalition as a Global Core Biodata Resource (GCBR). I am also preparing an application on behalf of current affiliation, RWTH Aachen University, to join the German Network for Bioinformatics Infrastructure (deNBI), which will include offering the Bioregistry as key bioinformatics infrastructure. Both applications require quantitative evidence of the usage and impact of the proposed resource as part of their respective key performance indicators (KPIs).
The Bioregistry by the Numbers
There are two kinds of statistics I think are important to convey about the Bioregistry. The first is related to community involvement. I’ve carefully planned the governance structure of the project to be sustainable (based on the open data, open code, and open infrastructure (O3) guidelines). This was successful in no small part due the way that there is a very low barrier for entry for small, external contributions (i.e., a drive-by curation). Therefore, it makes sense to highlight the number of unique contributors there have been to the data/code of the project as well as the volume of contributions in the forms of issues, discussions, and pull requests:
| Statistic | Count |
|---|---|
| Contributors | 75 |
| Open Issues | 146 |
| Closed Issues | 471 |
| Total Issues | 617 |
| Open pull requests | 13 |
| Closed pull requests | 1,012 |
| Total pull requests | 1,025 |
The second kind of statistic that’s important describes the content of the resource itself.
| Statistic | Count |
|---|---|
| Prefixes | 2,024 |
| Prefix Synonyms | 547 |
| External positive mappings | 9,162 |
| External negative mappings | 157 |
| Total external mappings | 9,319 |
| Collections | 18 |
| External Registries | 33 |
Note that this table is from mid-August 2025. Most of these numbers increase over time. To give some further context to these statistics, here’s a chart that shows how the Bioregistry stacks up against other related resources. Keep in mind, the Bioregistry is also a meta-resource that incorporates their important parts, too. You can understand this chart by looking at the percentage in parentheses (like +146% over Identifiers.org) and think: wow, those numbers are much bigger than 0%, which means the Bioregistry is much more comprehensive!
Direct Usage
Who uses the Bioregistry Website?
The Bioregistry website exposes metadata about ontologies, databases, and other resources that mint identifiers. This enables biocurators, data stewards, librarians, and others interested in identifying appropriate ontologies for reuse in their resources, data management plans, etc. It also allows researchers who consume linked (open) data to find context about the kinds of prefixes, CURIEs, or URIs that appear.
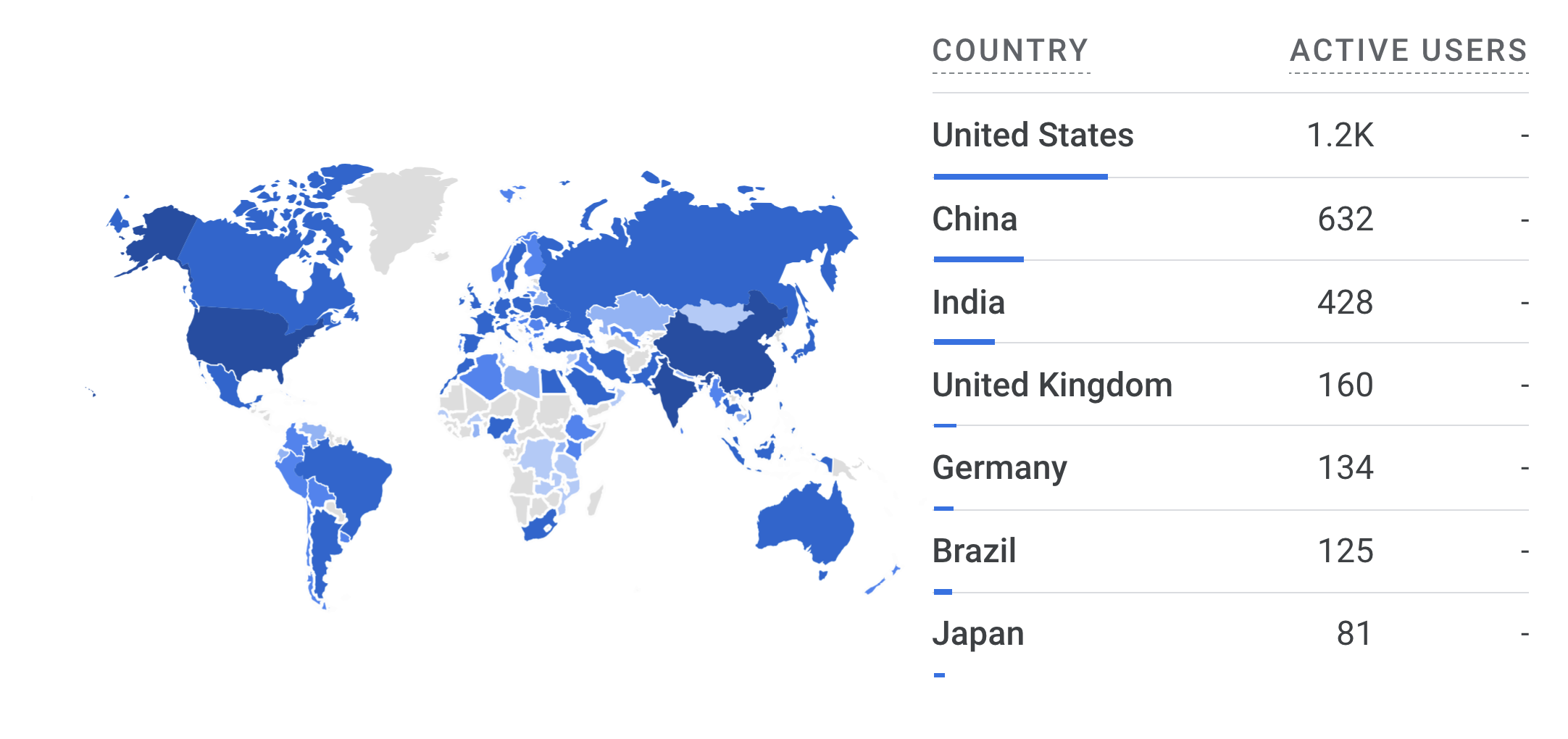
The following chart breaks down the nearly 4,400 unique users of the Bioregistry’s website by country. It shows that the Bioregistry is having a global impact, with potential for further growth in Africa (which I hope to do by getting in touch with the Africa PID Alliance).
API and Resolver Usage
Many usages of the Bioregistry are in the form of the resolution of links like
https://bioregistry.io/chembl:CHEMBL4303805.
Under the hood, the Bioregistry can expand prefixes like
chembl:CHEMBL4303805 to URLs.
This makes it a perfect service to support other applications that reference
entities and want to provide external links, without having to maintain the
links themselves.
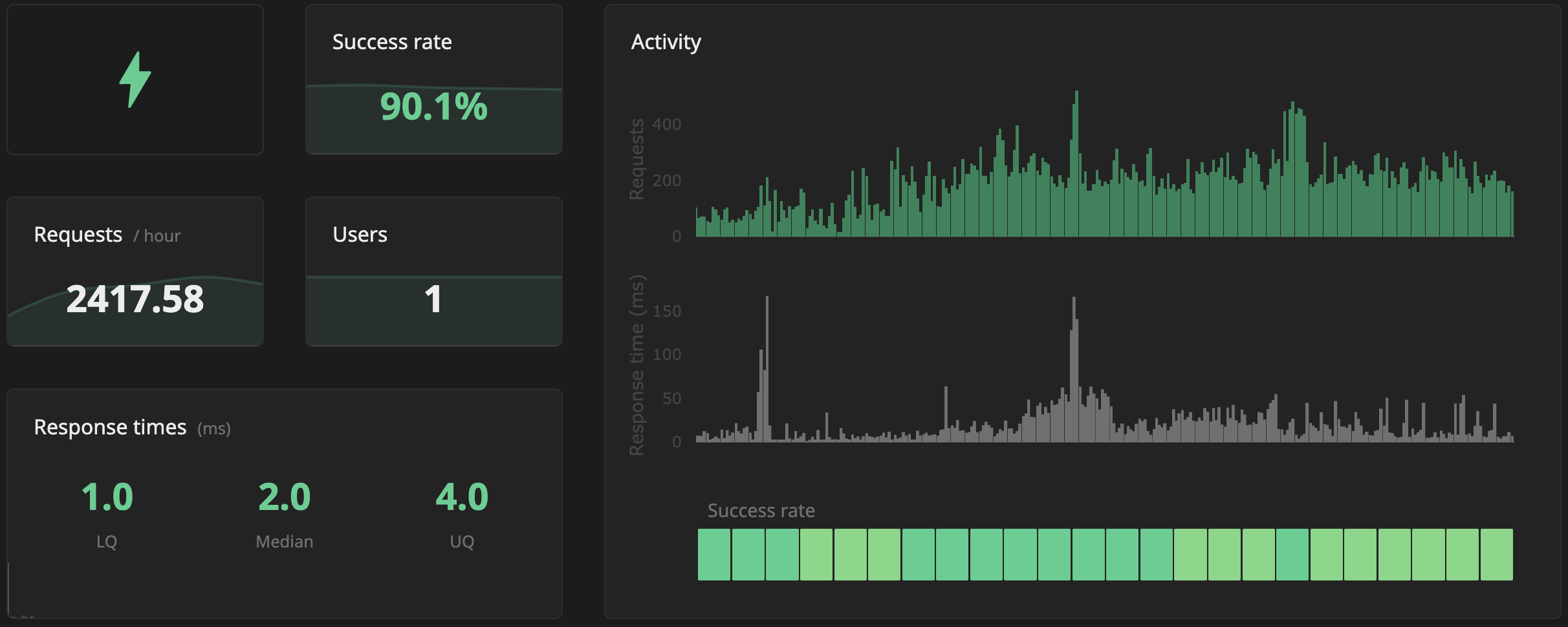
I’m just getting this working, so it should also be able to better keep track of unique users (note it’s just showing 1 so far) and their countries. Luckily, all of this is GDPR-compliant from the beginning.
Code Usage
The Bioregistry distributes a Python package that can be installed with
pip install bioregistry. I used
this query
to search GitHub for places where the bioregistry Python package is imported
and took detailed notes about their context (see details
here).
I found a large variety of usages across software packages, databases, ontologies, knowledge graphs, frameworks, and data models. Because the LinkML modeling language’s runtime indirectly depends on the Bioregistry, there are also dozens to hundreds of projects that indirectly use it. I also noted some usages from large organizations in the bioinformatics space, such as SciBite, Synapse, and the NFDI.
Indirect Usage
I used Wikidata as a backend to assess the indirect usage of the Bioregistry. I did this in a few steps:
- Make sure all direct usages have a Wikidata item that has a relationship to the Bioregistry Wikidata item via the uses P1547 (depends on software) or P2283 (uses) predicates.
- Identify well-known usages of direct usages, make sure they have Wikidata items, and are connected to them
- Automate querying Wikidata for all direct and indirect usages
- Search PubMed and quantify mentions of all direct and indirect usages
Indirect Software Dependencies
I’ve been developing quickstatements-client for automating adding content to Wikidata. I added an extension to it that pulls metadata from the Python Package Index (PyPI) and adds Python software packages. It does this recursively for a given package and its dependencies while adding appropriate P1547 (depends on software) relations between them.
Unfortunately, this workflow is still limited because it doesn’t find depedent software. This could be solved by doing a bulk download of PyPI and a large-scale network analysis. It might also be possible to extract this information from GitHub. However, for now, this is a good first step.
Indirect Ontology Dependencies via ODK
The Ontology Development Kit (ODK) uses the Bioregistry in several ways. It is used by many ontologies both in and out of the OBO Foundry. I developed a workflow for identifying ODK usage by searching GitHub and iteratively filtering out false positives. This resulted in over 140 repositories, more than half of which could directly be mapped back to the Bioregistry, given it tracks the repository associated with each prefix.
I have previously used quickstatements-client for automatically adding records for OBO Foundry ontologies. In next steps, I will extend this for the other ontologies identified by this analysis and also map them back to the ODK using the P1547 (depends on software) relation.
As a side note, for repositories using the ODK that couldn’t be mapped, I automated making stub curations in the Bioregistry, which lead to the curation of dozens of new prefixes.
Querying Wikidata
There’s a long tail of different ways to curate indirect usages of the Bioregistry. However, I believe that the largest cumulative impact for now will be though the lens of the ontologies built using it. After working through the scenarios above, I wrote a SPARQL query that recovers all direct and indirect dependencies (see live table below).
Literature Analysis
The final step was to analyze the literature for mentions of each of these direct and indirect dependencies. In typical Charlie fashion, I wanted to develop high-quality, modular, reusable software for doing this analysis, which lead to pubmed-downloader for wrapping API-based and bulk queries to PubMed (in addition to bulk downloading and processing, but that’s a story for a different blog post).
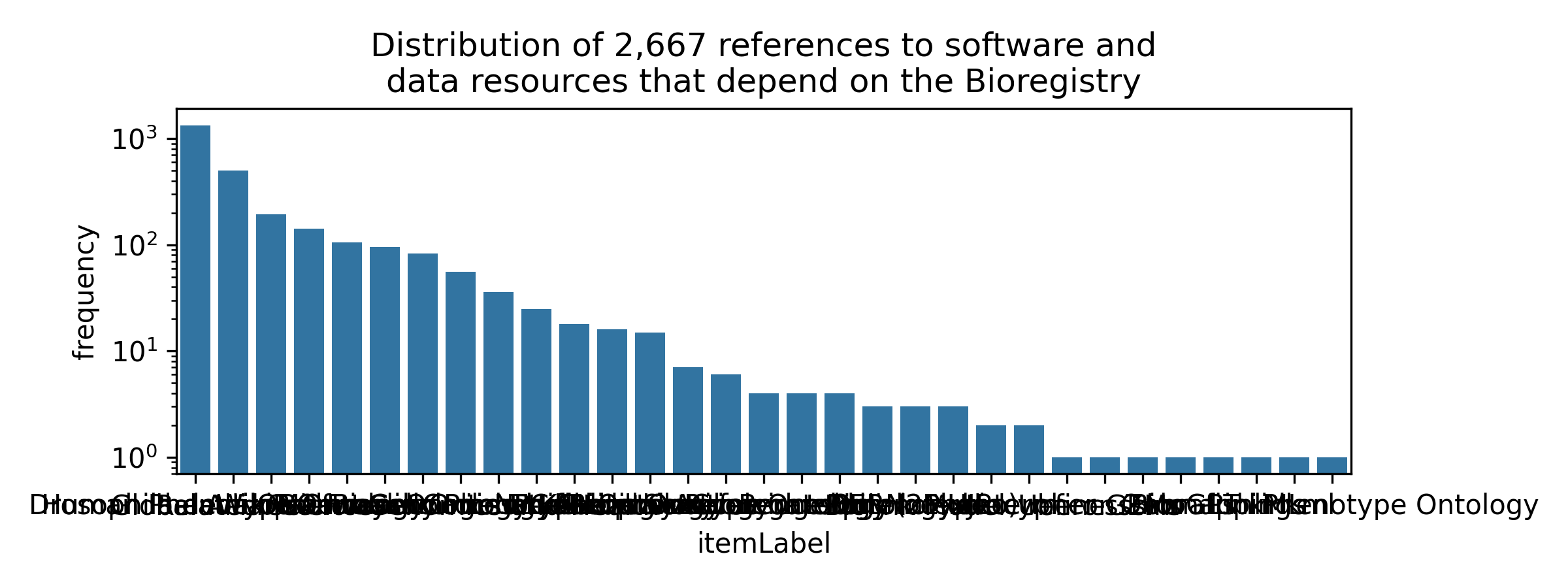
This is pretty great, it shows there are potentially thousands of papers that mention software, ontologies, databases, etc. that either directly or indirectly use the Bioregistry. It also shows, unsurprisingly, that there’s a power-law distribution in which is mentioned most in the literature.
The code for the literature analysis is available in its own repository. Of course, there is still lots of room for improvement and optimization, such as:
- Making the image above nicer!
- Using synonyms for search
- Removing false positives
- Incorporating citation networks
I’m interested to also make this work flow semi-automated to help assess the impact of other software/data resources, especially other key software that’s supporting the OBO Foundry like SSSOM-py.
Parting thoughts: it does seem like the Bioregistry is having a meaningful impact as a relatively young project. However, it’s still an uphill climb to get more direct recognition and adoption, which requires investing a lot of time in community building. This isn’t easy for me as an early career researcher not only because of lack of time/funding but also because of my relative lack of authority in the community of scientists who would benefit most from the Bioregistry (which comes with time).
I consider Chris Mungall and his group as the gold standard of being able to make large impact fast - they are known and trusted in the community, they have dedicated support staff that can focus on community management, they have many developers, and many projects where they can push their technologies. I’ll get there eventually!
