Exploring an unfamiliar SPARQL endpoint with the Bioregistry - a case study from NFDI4Culture
Earlier this week at the sixth NFDI4Chem consortium meeting, Torsten Schrade from the NFDI4Culture consortium gave a lovely and whimsical talk entitled A Data Alchemist’s Journey through NFDI which explored ways that we might federate and jointly query both consortia’s knowledge via their respective SPARQL endpoints. This post is about the very first steps I took when looking into this new (to me) SPARQL endpoint, namely to identify what prefixes and semantic spaces are present, then added a new CLI tool to the Bioregistry to do this reproducibly.
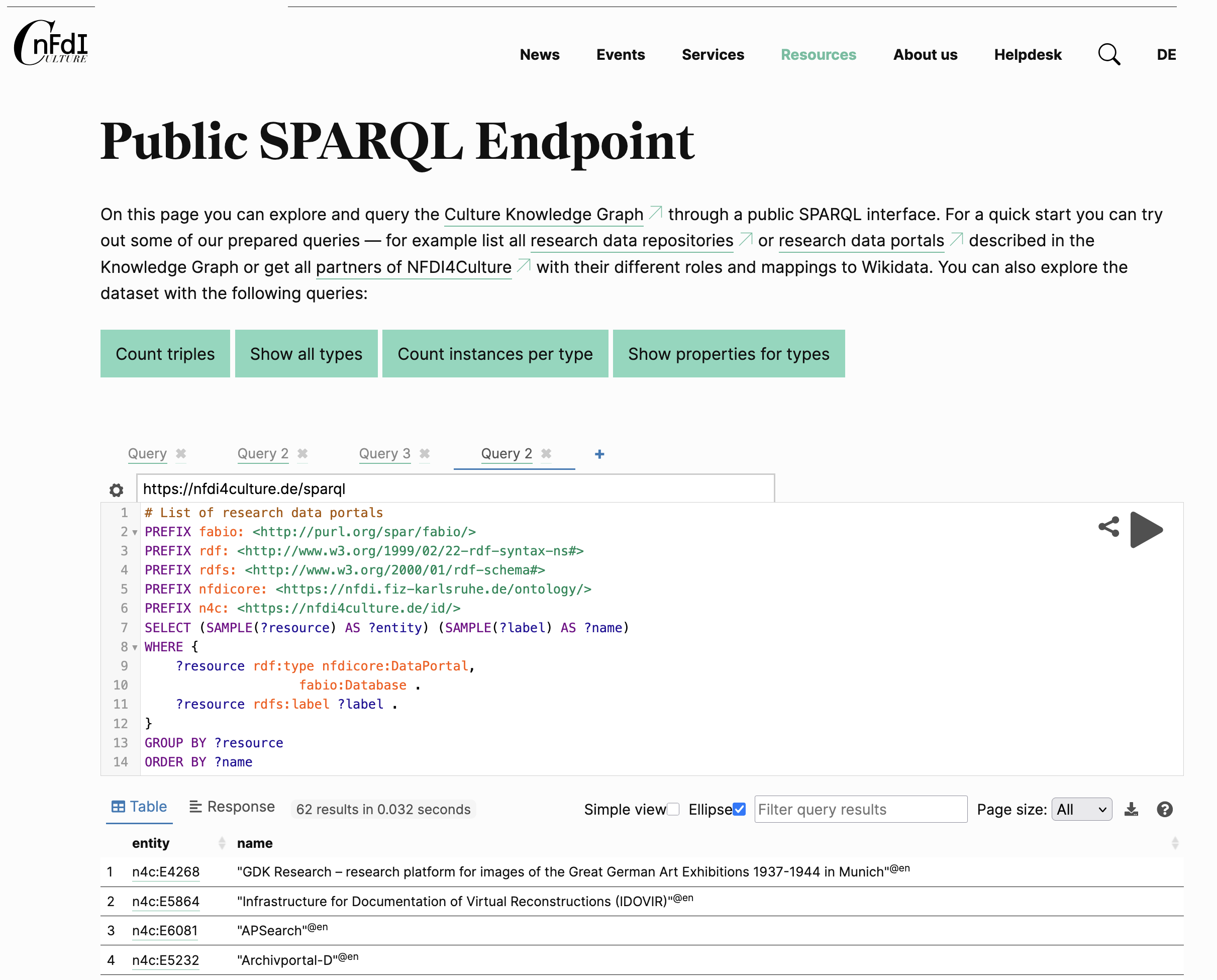
The NFDI4Culture’s SPARQL endpoint https://nfdi4culture.de/sparql is wrapped
by a nice user interface
here for interactive
querying in the browser.
It has two example queries to list all research data repositories and to list all research data portals, but otherwise I’m a bit stuck to better understand its schema.
Checking the Prefix Map
However, the NFDI4Culture SPARQL endpoint based on Virtuoso, so there is a way to look at what are the default CURIE prefixes and URI prefixes by navigating to here.
In my previous post, I demonstrated generalizing the notion of prefix map validation to incorporate prefix maps from either JSON-LD or in the beginning of turtle files.
I extended this even further to extract the prefix map from the Virtuoso SPARQL endpoint page in biopragmatics/bioregistry#1691. Note that this doesn’t work for all triple stores, it just works on Virtuoso because of the way that it provides a special page for showing the prefix map. I wasn’t able to find a way to get it directly, so the implementation does HTML scraping and parsing.
Here’s how you can use the validator I wrote:
$ bioregistry validate virtuoso https://nfdi4culture.de/sparql --tablefmt github
| prefix | uri_prefix | issue | solution |
|---|---|---|---|
| as | https://www.w3.org/ns/activitystreams# | unknown CURIE prefix | Switch to CURIE prefix ac, inferred from URI prefix |
| bif | http://www.openlinksw.com/schemas/bif# | unknown CURIE prefix | |
| dawgt | http://www.w3.org/2001/sw/DataAccess/tests/test-dawg# | unknown CURIE prefix | |
| dbpprop | http://dbpedia.org/property/ | unknown CURIE prefix | Switch to CURIE prefix dbpedia.property, inferred from URI prefix |
| fn | http://www.w3.org/2005/xpath-functions/# | unknown CURIE prefix | |
| formats | http://www.w3.org/ns/formats/ | unknown CURIE prefix | |
| gqi | http://www.openlinksw.com/schemas/graphql/intro# | unknown CURIE prefix | |
| gql | http://www.openlinksw.com/schemas/graphql# | unknown CURIE prefix | |
| gr | http://purl.org/goodrelations/v1# | unknown CURIE prefix | |
| ldp | http://www.w3.org/ns/ldp# | unknown CURIE prefix | |
| math | http://www.w3.org/2000/10/swap/math# | unknown CURIE prefix | |
| nci | http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl# | non-standard CURIE prefix | Switch to standard prefix: ncit |
| ogc | http://www.opengis.net/ | unknown CURIE prefix | |
| ogcgml | http://www.opengis.net/ont/gml# | unknown CURIE prefix | |
| ogcgs | http://www.opengis.net/ont/geosparql# | unknown CURIE prefix | |
| ogcgsf | http://www.opengis.net/def/function/geosparql/ | unknown CURIE prefix | |
| ogcgsr | http://www.opengis.net/def/rule/geosparql/ | unknown CURIE prefix | |
| ogcsf | http://www.opengis.net/ont/sf# | unknown CURIE prefix | |
| product | http://www.buy.com/rss/module/productV2/ | unknown CURIE prefix | |
| protseq | http://purl.org/science/protein/bysequence/ | unknown CURIE prefix | |
| rdfdf | http://www.openlinksw.com/virtrdf-data-formats# | unknown CURIE prefix | |
| sc | http://purl.org/science/owl/sciencecommons/ | unknown CURIE prefix | |
| scovo | http://purl.org/NET/scovo# | unknown CURIE prefix | |
| sd | http://www.w3.org/ns/sparql-service-description# | unknown CURIE prefix | |
| sioc | http://rdfs.org/sioc/ns# | unknown CURIE prefix | Switch to CURIE prefix sioc.core, inferred from URI prefix |
| sql | http://www.openlinksw.com/schemas/sql# | unknown CURIE prefix | |
| stat | http://www.w3.org/ns/posix/stat# | unknown CURIE prefix | |
| vcard2006 | http://www.w3.org/2006/vcard/ns# | unknown CURIE prefix | Switch to CURIE prefix vcard, inferred from URI prefix |
| virtcxml | http://www.openlinksw.com/schemas/virtcxml# | unknown CURIE prefix | |
| virtrdf | http://www.openlinksw.com/schemas/virtrdf# | unknown CURIE prefix | |
| xf | http://www.w3.org/2004/07/xpath-functions | unknown CURIE prefix | |
| xsl10 | http://www.w3.org/XSL/Transform/1.0 | unknown CURIE prefix | |
| xsl1999 | http://www.w3.org/1999/XSL/Transform | unknown CURIE prefix | |
| xslwd | http://www.w3.org/TR/WD-xsl | unknown CURIE prefix | |
| yago | http://dbpedia.org/class/yago/ | unknown CURIE prefix |
Interpreting the results
Some of the key takeaways from this table are:
- The feedback on
asis a false positive - a look at https://www.w3.org/ns/activitystreams# shows that the W3 standard wantsasto be the preferred prefix - There are several true positive suggestions, like fixing the
ncitprefix. - There’s a whole group of URI spaces using
opengis.netfrom the Open Geospatial Consortium, dealing with geospatial data that could be registered in the Bioregistry - There are a large number URI spaces from
openlinksw.com, which correspond to Virtuoso itself. These might be good to put in the Bioregistry, but are all very short, which means that there is high potential for overlap. However, there aren’t any reported conflicts - There are several
w3.orgstandard prefixes that should be registered in the Bioregistry. - There are several prefixes that I’m not familiar with that also use short acronyms, which makes me a bit hesitant to add in the Bioregistry. In these cases, I usually either don’t add them because of lack of wide-spread use or add them using a less generic prefix.
Something I still want to implement is a generic workflow for identifying putative URI spaces in a given remote SPARQL endpoint. If there are only a small number of triples that can be exhaustively queried, then this workflow can be used. Otherwise, I am still considering different options.