Machine-Actionable Training Materials at BioHackathon Germany 2025
I recently attended the 4th BioHackathon Germany hosted by the German Network for Bioinformatics Infrastructure (de.NBI). I participated in the project On the Path to Machine-actionable Training Materials in order to improve the interoperability between DALIA, TeSS, mTeSS-X, and Schema.org. This post gives a summary of the activities leading up to the hackathon and the results of our happy hacking.
Team

Our project, On the Path to Machine-actionable Training Materials, had the following active participants throughout the week:
- Nick Juty & Phil Reed (University of Manchester)
- Leyla Jael Castro & Roman Baum (Deutsche Zentralbibliothek für Medizin; ZB Med)
- Petra Steiner (University of Darmstadt)
- Oliver Knodel & Martin Voigt (Helmholtz-Zentrum Dresden-Rossendorf; HZDR)
- Dilfuza Djamalova (Forschungszentrum Jülich; FZJ)
- Jacobo Miranda (European Molecular Biology Laboratory; EMBL)
Nick and Petra were our team leaders and Phil acted as the project’s de facto secretary. On the first day of the hackathon, we were briefly joined by Alban Gaignard (Nantes University), Dimitris Panouris (SciLifeLab), and Harshita Gupta (SciLifeLab) to present their current related work. Similarly, Dominik Brilhaus (Heinrich-Heine-Universität Düsseldorf) joined on the first day to share his perspective from DataPLANT (the NFDI consortium for plants) as a training materials creator. Finally, Helena Schnitzer (FZJ) participated in some Schema.org discussions through the week.
Goals
We categorized our work plan into three streams:
- Training Material Interoperability - survey the landscape of relevant ontologies and schemas for annotating learning materials, curate mappings/crosswalks between existing data models, develop a programmatic toolbox, and begin federating between training material platforms
- Training Material Analysis - analyze training materials at scale to group similar training materials, reduce redundancy, and semi-automatically construct learning paths
- Modeling Learning Paths - collect use cases and develop a (meta)data model for learning paths
Training Material Interoperability
Interoperability is third pillar of the FAIR data principles. Metadata describing training materials may be captured and stored in one of several data models including the DALIA Interchange Format (DIF) v1.3, the format implicitly defined by the TeSS API, and the Schemas.org Learning Material profile. Further, metadata records conforming to these data models are filled with references to terms in other ontologies, controlled vocabularies, databases, and other resources that mint (persistent) identifiers. Our overarching goal at the hackathon was to improve interoperability on both levels.
Indexing Ontologies and Schemas
Our first concrete goal for training material interoperability at the hackathon was to survey ontologies, controlled vocabularies, databases, and other resources that mint (persistent) identifiers that might appear in the metadata describing a learning material. For example, TeSS uses the EDAM Ontology to annotate topics onto training materials. For the same purpose, DALIA uses the Hochschulcampus Ressourcentypen (I’ll say more on how we deal with the conflicting resources in the section below on mappings).
Our second concrete goal was to survey schemas that are used in modeling open educational resources and training materials, for example, Schema.org, OERSchema, and MoDALIA, which encodes the DALIA Interchange Format (DIF) v1.3.
The Semantic Farm (https://semantic.farm) is comprehensive database of metadata about resources that mint (persistent) identifiers (e.g., ontologies, controlled vocabularies, databases, schemas) such as their preferred CURIE prefix for usage in SPARQL queries and other semantic web applications. It imports and aligns with other databases like Identifiers.org (for the life sciences) and BARTOC (for the digital humanities) to support interoperability and sustainability. It follows the open data, open code, and open infrastructure (O3) guidelines and has well-defined governance to enable community maintenance and support longevity.
It’s the perfect place to index all the learning material and open educational resource-related ontologies, controlled vocabularies, databases, and schemas.
I gave a tutorial on how to search the Semantic Farm for ontologies, controlled vocabularies and other resources that mint (persistent) identifiers, and how to contribute any that are missing. In short, they can be contributed by filling out the new prefix request template on GitHub. If you’re interested to add a new entry, you can directly use the form, read the contribution guidelines, or watch a short YouTube tutorial.
While I had done some significant preparatory work before the hackathon by creating many new entries in the Semantic Farm, the team found and added several new and important entries to the Semantic Farm during the hackathon too. Here are two highlights:
Martin Voigt contributed the prefix
amb for the
Allgemeines Metadatenprofil für Bildungsressourcen
(General Metadata Profile for Educational Resources) in
biopragmatics/bioregistry#1781.
This is a metadata schema for learning materials produced by the
Kompetenzzentrum Interoperable Metadaten (KIM) within the Deutsche Initiative
für Netzwerkinformation e.V. that was heavily inspired by
Schema.org and the Dublin Core
Learning Resource Metadata Initiative (LRMI)
Dilfuza Djamalova and
Jacobo Miranda contributed the prefix
gtn for
Galaxy Training Network
training materials in
biopragmatics/bioregistry#1779.
This resource contains multi- and cross-disciplinary training materials for
using the Galaxy workflow management system. Below, I describe how we ingested
transformed the training materials from GTN into a common format such they can
be represented according to the DALIA Interchange Format (DIF) v1.3, the
implicit data model expected by TeSS, and in Schema.org-compliant RDF.
Ultimately, we collated relevant ontologies, controlled vocabularies, schemas and other resources that mint (persistent) identifiers in a collection such that they can be easily found and shared.
Semantic Mappings and Crosswalks
I alluded to the different resources used by TeSS and DALIA to annotate disciplines. The issue of partially overlapping ontologies, controlled vocabularies, and database is quite widespread, and can manifest in a few different ways. The figure above shows that redundancy can arise because of different focus within a domain (i.e., the chemistry example), different hierarchical specificity (i.e., the disease example), and due to massive generic resources having overlap across many domains (e.g., like UMLS, MeSH, NCIT).
This is problematic when integrating learning materials from different sources, e.g., TeSS and DALIA, because two learning materials may be annotated with different terms describing the same discipline. Therefore, the solution is to create semantic mappings between these terms.
I’ve worked for several years on the Simple Standard for Sharing Ontological Mappings (SSSOM) standard for storing semantic mappings, so this was naturally the target for our work. Further, I have been working on a domain-agnostic workflow for predicting semantic mappings with lexical matching and deploying a curation interface called SSSOM Curator. I gave a tutorial for using SSSOM Curator to the team based on a previous tutorial I made (that can be found on YouTube here). We prepared predicted semantic mappings between several learning material-related ontologies in biopragmatics/biomappings#204, but we didn’t prioritize semantic mapping curation during the hackathon. Here’s what they look like in the SSSOM Curator interface for Biomappings:
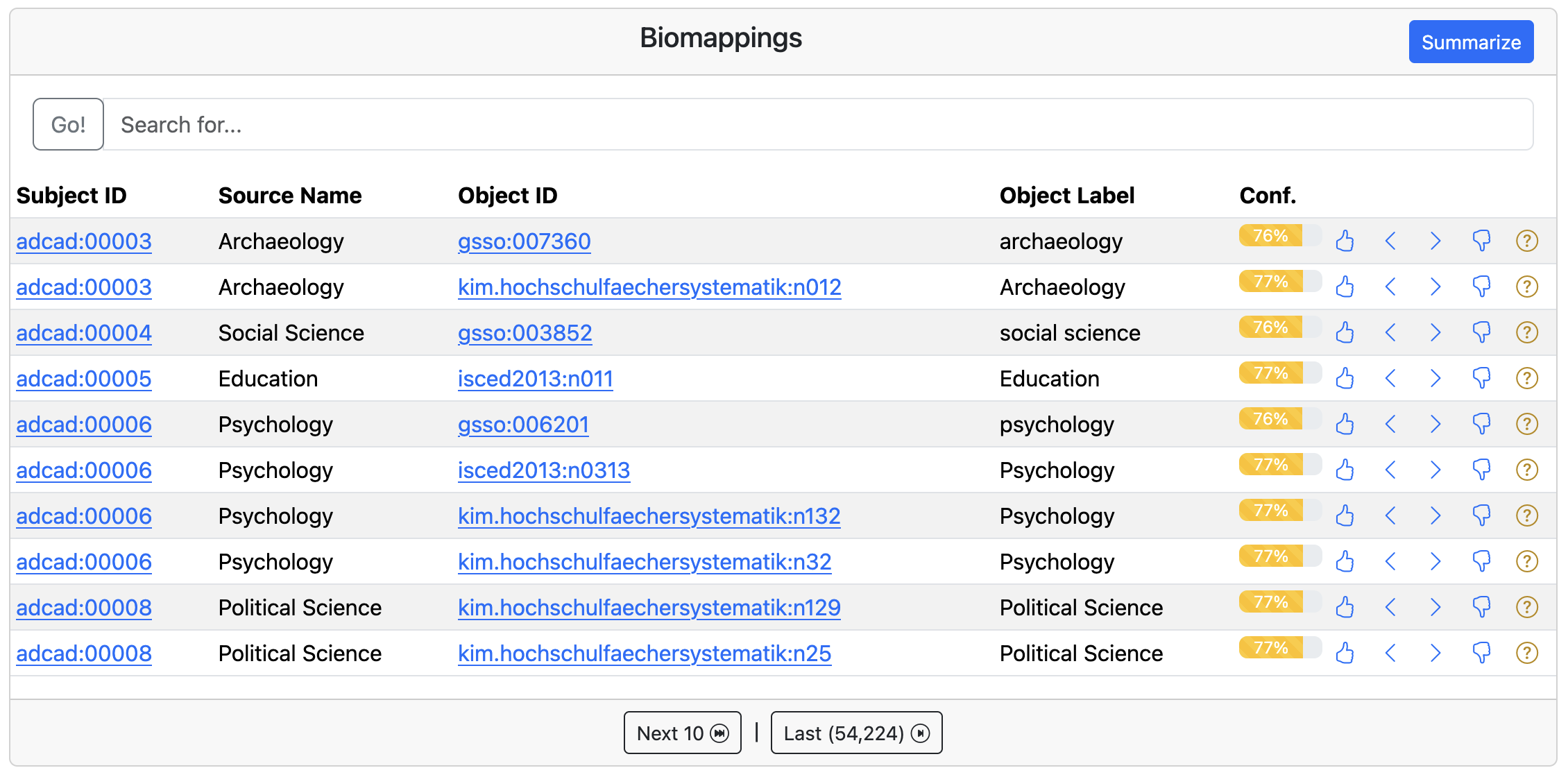
Where curating correspondences between concepts in ontology, controlled vocabularies, and databases is often called semantic mapping, curating correspondences between schemas and properties therein is often called crosswalks. We put a bigger emphasis on producing crosswalks between Schema.org and MoDALIA. This is actually a more complex problem due to the fact that correspondences between elements in schemas can be more sophisticated (e.g., mapping between two fields for first and last names to a single name field), but there are at least a few places where properties can be mapped with SSSOM.
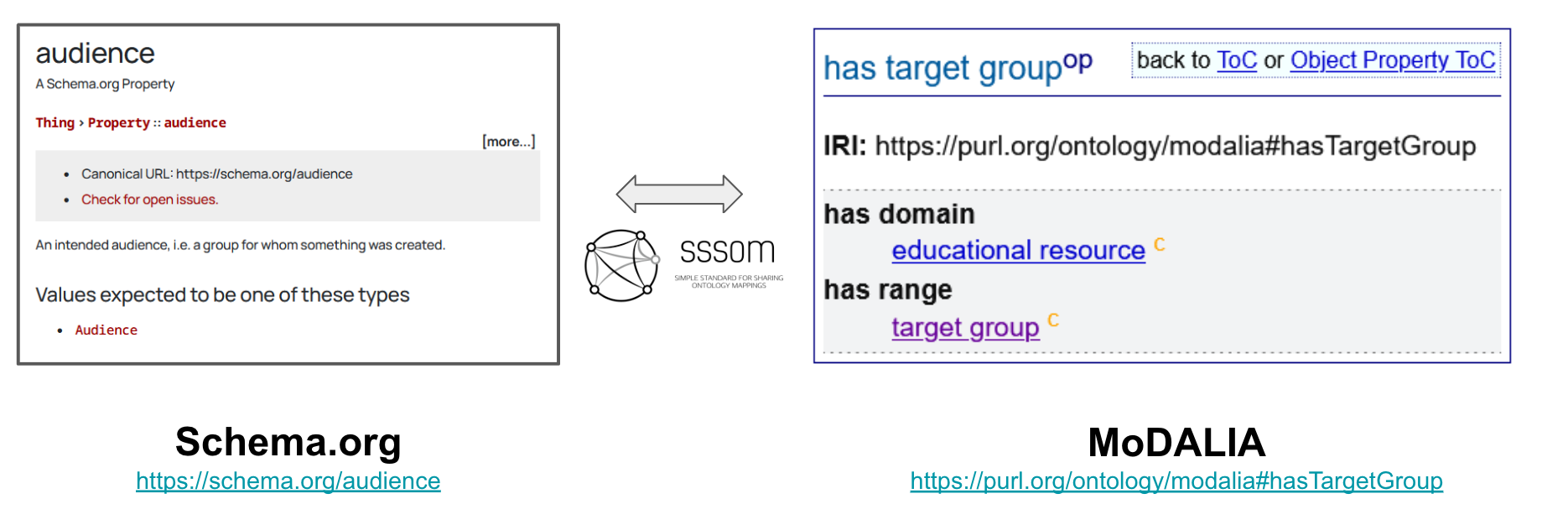
An interesting lesson learned is that some curators find using SKOS
relationships challenging because the narrow and broader relations have the
opposite direction than what they would expect. For example,
X skos:narrowMatch Y means that X is narrower than Y, not X has a narrow match
Y. Many vocabularies use a verb as part of the predicate to reduce this
confusion - I’m sure if it were X skos:isNarrowMatchFor Y, then this would not
have been a problem. Deep down, the real issue is that transparent identifiers
(i.e., human-readable ones) are bad, since they can’t be changed over time. See
the excellent article,
Identifiers for the 21st century,
by McMurry et al. (2017) for a more detailed discussion on what makes a good
identifier.
Operationalizing Crosswalks
The next step was to translate the abstract crosswalks between DALIA, TeSS, and Schema.org into a concrete implementation using a general purpose programming language (i.e., Python).
The Scaling Problem
Given that we only focused on these three data models, it’s not unrealistic to produce a DALIA-TeSS crosswalk, TeSS-Schema.org crosswalk, and DALIA-Schema.org crosswalk. However, this approach does not scale well - in general, it requires curating and implementing ${N}\choose{2}$ crosswalks with $N$ being the number of schemas.
An alternative is to use a hub-and-spoke model, in which one data model is targeted as the intermediary used for interchange and storage. This reduces the burden on curators of crosswalks, as they only have to curate a single crosswalk for any given data model into the intermediary. Similarly, it reduces the burden on code maintainers as only a single crosswalk has to be implemented.
The challenge with open educational resources and learning materials is that no existing data model is sufficient to cover the (most important) aspects of all other data models. This motivated us to implement a unified, generic data model for learning materials to serve as the interoperability hub between DALIA, TeSS, Schema.org, and other data models.
graph TD
subgraph alltoall ["All-to-All (complex, burdensome)"]
dalia[DALIA] <--> tess[TeSS]
dalia <--> schema[Schema.org]
dalia <--> oerschema[OERschema]
dalia <--> amb["Allgemeines Metadatenprofil für Bildungsressourcen (AMB)"]
dalia <--> lrmi["Learning Resource Metadata Initiative (LRMI)"]
dalia <--> erudite[ERuDIte]
tess <--> schema
tess <--> oerschema
tess <--> amb
tess <--> lrmi
tess <--> erudite
schema <--> oerschema
schema <--> amb
schema <--> lrmi
schema <--> erudite
oerschema <--> amb
oerschema <--> lrmi
oerschema <--> erudite
amb <--> lrmi
amb <--> erudite
lrmi <--> erudite
end
subgraph hub ["Hub-and-Spoke (maintainable, extensible)"]
direction TB
hubn[Unified OER Data Model] <--> daliaspoke[DALIA]
hubn[Unified OER Data Model] <--> tessspoke[TeSS]
hubn[Unified OER Data Model] <--> schemaspoke[Schema.org]
hubn[Unified OER Data Model] <--> oerschemaspoke[OERschema]
hubn[Unified OER Data Model] <--> ambspoke["Allgemeines Metadatenprofil für Bildungsressourcen (AMB)"]
hubn[Unified OER Data Model] <--> lrmispoke["Learning Resource Metadata Initiative (LRMI)"]
hubn[Unified OER Data Model] <--> eruditespoke[ERuDIte]
end
alltoall --> hub
The famous XKCD comic, Standards (https://xkcd.com/927), proselytizes that any proposal of a unified standard that covers everyone’s use cases is doomed to be an $N+1$ competing standard. While I’m doing my best to present the work done in preparation for the hackathon and at the hackathon in a linear way, the truth is that most steps also included discussion, hacking, trying, failing, and repeating. Therefore, I can confidently say that for practical reasons, implementing a new de facto standard was the only realistic choice.
The OERbservatory Data Model

During the hackathon, we implemented the open source OERbservatory Python package. I first want to talk about three major features that it includes:
- a unified, generic object model for open educational resources that’s effectively the union of the best parts of DALIA, TeSS, Schema.org, and a few other data models we found
- import and export to two open educational resource and learning materials data models - DALIA and TeSS. We didn’t have time during the hackathon to implement import and export to Schema.org.
- import from three external learning material repositories - OERhub, OERSI, and the Galaxy Training Network (GTN)
Here’s an excerpt of the object model, implemented using Pydantic. Note that Pydantic uses a combination of Python’s type system and type annotations to express constraints and rules, similarly to how SHACL does. However, we get the benefit of Python type checking and the Python runtime to check that we’ve encoded this all correctly. Finally, all Pydantic models can be serialized and deserialized from JSON.
class EducationalResource(BaseModel):
"""Represents an educational resource."""
model_config = ConfigDict(arbitrary_types_allowed=True)
reference: Reference | None = Field(
None,
description="The primary reference for this learning material",
examples=[Reference(prefix="dalia", identifier="")]
)
title: InternationalizedStr = Field(..., description="The title of the learning material")
authors: list[Author | Organization] = Field(
default_factory=list,
description="An ordered list of authors (i.e., persons or organizations) of the learning material",
examples=[
Author(name="Charles Tapley Hoyt", orcid="0000-0003-4423-4370"),
Organization(name="NFDI", ror="05qj6w324"),
],
min_len=1,
)
...
Technology Comparison (content warning: programming culture wars)
DALIA and Schema.org built on top of semantic web principles. Records about learning materials encoded in these data models are stored in RDF and queryable via SPARQL. However, while powerful, SPARQL is a querying language that is inherently limited in its expressibility and utility. A general purpose programming language is more suited for building data science workflows, search engines, APIs, web interfaces, and other tools on top of open educational resource and learning material data. That's why we emphasized concretizing the crosswalks between DALIA, TeSS, and Schema.org in a software implementation.
We chose Python as the target language because of its ubiquity and ease of use. When the TeSS platform was initially developed in the early 2010s, the Ruby programming language and the Ruby on Rails framework were a popular choice for developing web applications. Unfortunately, the scientific Python stack and machine learning ecosystem led Python to being a clear winner for academics and scientists. This creates an issue that only a small number of academics are skilled in Ruby and can participate in the development of TeSS.
It was also crucial that we used Python such that our implementation was reusable. For example, the DALIA 1.0 platform was implemented using Django, which made it effectively impossible to reuse any of the underlying code outside, e.g., in a data science workflow. The same issue is also true for the TeSS implementation using Ruby-on-Rails. While these batteries-included frameworks can get a minimal web application running quickly, they generally lead developers towards writing code that isn't reusable.
OERbservatory as an Interoperability Hub between DALIA and TeSS
Before we even started working on the OERbservatory, we had implemented two packages for working with data in DALIA and TeSS:
- data-literacy-alliance/dalia-dif implements a parser for the DALIA DIF v1.3 tabular format, an internal representation of the content (also using Pydantic), and an RDF serializer (using on pydantic-metamodel
- cthoyt/tess-downloader implements an API client to TeSS and an internal representation of the learning resource data model (using Pydantic)
Because each of these packages already implemented an internal (lossless) representations of the data models for DALIA and TeSS, respectively, we only had to write code in the OERbservatory that mapped the fields between them to OERbservatory’s data model.
This was a big milestone towards interoperability. We demonstrated its potential by programmatically downloading all learning materials from the ELIXIR TeSS instance’s API and exporting them as DALIA RDF. Similarly, we converted all learning materials curated for DALIA into the TeSS JSON format. Later, I’ll describe how we took this workflow one step further to implement syncing between DALIA and TeSS.
Note that this mapping can’t simply be expressed using SSSOM, SHACL, or other declarative languages, because it relies on more sophisticated logic. For example, topics annotated with ontology terms in the DALIA data model only store the URI reference, whereas topics annotated with ontology terms in the TeSS data model require both the URI reference and the term’s label. Since we’re encoding our crosswalks using a general purpose programming language, we have a larger toolkit available. Here, we could use PyOBO, a generic package I’ve written for working with ontologies, for looking up labels.
Unfortunately, we did not have time to implement an importer/exporter for
Schema.org. We deprioritized this because Schema.org it felt the least
approachable due to the way its documentation is written, the complexity of its
models, and prolific use of mixins. We considered if we could automatically
generate Pydantic classes from Schema.org - and it turns out that
pydantic-schemaorg has
already done it! Unfortunately, the code is not compatible with modern versions
of Pydantic, and the project appears abandoned. We only had so much time at the
hackathon, so forking/reviving/rewriting pydantic-schemaorg was left as a task
for later.
The OERbservatory as an Aggregator
Besides open educational resources and learning materials that are encoded in
the DALIA, TeSS, and Schema.org formats, there are many repositories of learning
materials that do not conform to a well-defined schema. Prior to the hackathon,
I had already explored the Austrian OERhub and
Open Educational Resources Search Index (OERSI)
and written importers into dalia-dif. At the hackathon, I reimplemented those
importers using the newly formed OERbservatory unified, generic data model.
On the Thursday morning of the BioHackathon, I had an excellent mob programming session with Dilfuza Djamalova and Jacobo Miranda to import training materials from the Galaxy Training Network (GTN). It turns out that there are already several open educational resources and learning materials that are automatically scraped and imported by TeSS. However, those importers are limited by TeSS’s relatively rigid data model, which is bound to their database and can therefore not be easily evolved. Dilfuza and Jacobo had a few goals for our hacking:
- There are fields in GTN that aren’t yet captured by TeSS. They wanted to implement those fields in OERbservatory, demonstrate their usage, then gently nudge TeSS to evolve its data model to support their use cases
- They wanted to index their content in DALIA, which becomes much easier if they only have to maintain one importer in OERbservatory which can already export to DALIA
- GTN is part of the DeKCD consortia, which wants to deduplicate training material. Adding an importer here gives access to the workflows we’re building for reconciling different metadata curated in different places about the same materials, and identifying similar materials to reduce duplicate effort, and connect people working on the same kinds of materials
We implemented the GTN importer in data-literacy-alliance/oerbservatory#8 which covers tutorials in GTN and later could be extended to slide decks. Along the way, we updated the main educational resource model in OERbservatory to include a few new fields, including status (which also is shared by TeSS- that now needs to be incorporated), the publication date, and the modified date. We did not make a complete mapping for all fields in GTN due to time constraints, so we implemented logging that summarizes fields that haven’t yet been mapped (see the PR for examples of each). For example, the way that contributor information is incorporated into the API from the frontmatter in the source is interesting - it resolves the keys in the frontmatter to entries in this YAML file in the GTN GitHub repository. We will want to think about the best way to map the authors into OERbservatory, and this also might be a time to extend the author list to include contributor role annotations.
I was very excited that Dilfuza and Jacobo were motivated to work on this and contribute following the hackathon. We see if the OERbservatory is approachable enough for future external contributions! For example, Robert Hasse of NFDI4BIOIMAGE already proactively prepared a script that exports their consortium’s training materials into the DALIA DIF v1.3 tabular format. I don’t consider this a very approachable format, and I’m sure efforts like his could have been eased by using OERbservatory as a target. The next steps are to incorporate the Swiss Digital Research Infrastructure for Arts and Humanities (DARIAH-CH) and Physical Sciences Data Infrastructure (PSDI) learning materials, which appeared on the schematic diagram for OERbservatory earlier. There are also a lot of other potential learning material repositories to scrape like Glittr.com. If you have a suggestion, you can drop it in the OERbservatory issue tracker. Further, given that Martin Voigt was in the room during this hacking and discussion, and he is the maintainer for TeSS’s scraper code, we already started formulating plans on how we might be able to deduplicate efforts.
Federation of Open Educational Resources and Learning Materials
The next step towards interoperability beyond the demonstration of converting between formats used by DALIA and TeSS was to demonstrate actually posting the content to the live services.
While we are currently in the process of implementing submission of open educational resources and learning materials in DALIA, TeSS already has a web-based interface for registering new learning materials. TeSS doesn’t have a documented API endpoint for posting learning materials, but luckily, Martin knew where it was and helped to figure out the correct way to pass credentials to use it. We managed this by a combination of reading the Ruby implementation of TeSS and good ‘ol trial and error. In the end, we implemented posting learning materials in the TeSS-specific Python package in cthoyt/tess-downloader#2. Then, it was only a matter of stringing together code that converts DALIA to OERbservatory, OERbservatory to TeSS, and then to upload to TeSS.
In parallel, Martin worked on improving the devops behind the PaNOSC TeSSHub to enable quicky spinning up new TeSS instances that each have their own subdomain. He created a different subdomain for each of DALIA, OERSI, GTN/KCD, and OERhub. Finally, we wrote a script that uploaded all open educational resources and learning material from each source to the appropriate TeSS instance in data-literacy-alliance/oerbservatory#3. The results in each space can be explored here:
| Source | Domain |
|---|---|
| DALIA | https://dalia.tesshub.hzdr.de |
| OERhub | https://oerhub.tesshub.hzdr.de |
| OERSI | https://oersi.tesshub.hzdr.de |
| GTN/deKCD | https://kcd.tesshub.hzdr.de |
| PanOSC | https://panosc.tesshub.hzdr.de |
A full list of spaces can be found here.
European Open Science Cloud
The great specter looming over most NFDI-related projects is how to interface with the European Open Science Cloud (EOSC). At the surface, EOSC is a massive undertaking to democratize access to research infrastructure on the European level. However, having just entered the NFDI bubble at the end of the summer, I have bene overwhelmed by the high pressure to participate in EOSC combine with the lack of funding and lack of direction on how to best go about doing that. All of that being said, Oliver Knödel spend the hackathon preparing the concept for how we could connect TeSSHub to the EOSC open educational resource and training materials registry using the Open Archives Initiative Protocol for Metadata Harvesting (OAI-PMH). Once TeSSHub can demonstrate federating its content through this mechanism, we can use as inspiration to make a generic implementation in OERbservatory.
Governance and Provenance
Now that it’s possible to copy training materials from one platform to another, we have started to consider governance and provenance issues like:
- If a training material originally curated in DALIA is displayed in TeSS, how is that attributed? We will have to carefully consider how metadata records about learning resources are identified, and how those identifiers are passed around during interchange/syncing.
- If a training material originally from TeSS is enriched in the DALIA platform, should that information flow back to TeSS, and how? We will have to carefully consider how information is deduplicated and reconciled
- How do we implement technical systems that can keep many federated platforms up-to-date with each other?
I’m sure there will be many more questions along these lines. Luckily, the mTeSS-X group has already begun discussions on a smaller scale, since they care about how to federate between many disparate TeSS instances.
Training Material Analysis
Our team split into two for the analysis of training materials. The first team looked into algorithmic mechanisms for featurizing open educational resources and learning materials and applications of those features. The second team looked into using large language models (LLMs) for the automated construction of learning paths.
Featurization and Application
The first team looked into two techniques for featurizing (i.e., assigning dense vectors) to open educational resources and learning materials.
The first and most interpretable technique was to concatenate free text fields and labels from structured fields from a learning resource and index the entire corpus (i.e., all learning resources) using the term frequency-inverse document frequency (TF-IDF) algorithm. This does a small amount of text preprocessing, calculates a word list for the entire corpus, then calculates for each word the likelihood of appearance in a given learning material versus the entire corpus. Then, each learning material is assigned a dense vector with values from $[0, 1]$ the length of the word list. Learning materials can be compared, e.g., using cosine similarity between their respective vectors.
The second technique was to use the sentence transformers machine learning architecture, which relies on a pre-trained (not large) language model to accomplish a similar vectorization. Both methods run in less than a few minutes for the corpus of learning resources from DALIA, TeSS, OERHub, and OERSI. We also pre-calculated the all-by-all similarities and applied a cutoff of 0.7 to shorten the list. Both the TF-IDF and sentence transformers vector index and similarities are commit to the OERbservatory repository and are available here.
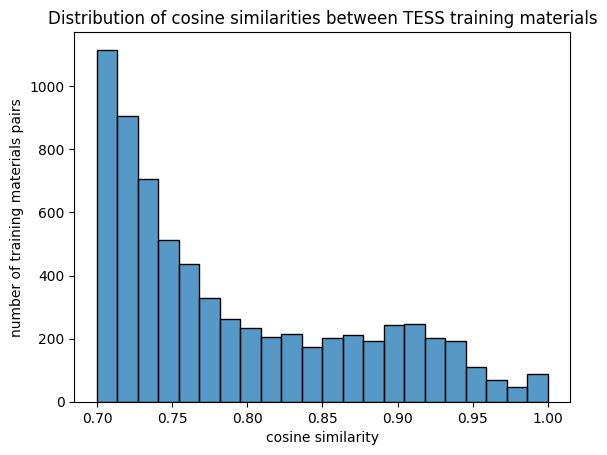
After we had embeddings, Dilfuza began to investigate some of the following:
- Identify duplicates metadata records corresponding to the same learning material resource, e.g., when two different platforms scraped the same learning material
- Semi-automatically identify similar training materials both to improve suggestions to learners, to connect the learning material creators, and to help de-duplicate training material creation efforts
We only managed to get this far in the last day of the hackathon, so there is still a lot more to do here! Originally, I had planned on also using these embeddings to train classifiers for key provenance metadata such as topic, target audience, and difficulty level, then to create a semi-automated curation workflow for enriching learning materials whose records were sparse with annotation. These will be next steps.
Automated Construction of Learning Paths
Nick looked into using large language models (LLMs) to construct learning paths through machine-assisted dialog. This part is highly experimental so there isn’t much to point to yet, but the idea was to take in a list of learning materials (either hard-coded or as a URL for the chat system to retrieve) and a prompt to ask the LLM ot collect similar materials base don objectives and keywords, then create a learning path based on difficult (which is infrequently annotated) and suggest a title.
This workflow was used to produce three learning paths on the following topics that were each ordered, had reference links, a difficulty rating, a title, and provider:
- Sequencing and QC (10 items)
- Git and Version Control (6 items)
- Genome Annotation (8 items)
More on this in future work!
Modeling Learning Paths
While there isn’t a clear consensus on what a learning path is, a simple definition is that a learning path is a sequence of learning materials to consume to help a learner achieve a specific level of competence on a topic. TeSS implements a data model for learning paths based on this definition and the ELIXIR TeSS instance has eleven examples. Our team had the goal to develop an extension Schemas.org (in Bioschemas) to capture learning paths.
For transparency, I didn’t actively participate in this track, but think it’s worth sharing the results, most of which are adapted from Phil’s repository in BioSchemas/LearningPath-sandbox.
Proposed Data Model
Phil, Alban, and Leyla proposed two new Bioschemas profiles and a small change to one Bioschemas profile with the help of Nick and Roman:
LearningPath: inherits fromCourseLearningPathModule: inherits fromCourse,Syllabus,ListItem, andItemListTrainingMaterial: inherits fromLearningResourceandListItem
Here’s a class diagram describing the proposed data model, where 🔺 is Schema.org type, 🟩 is Bioschemas profile, 🔵 is new profile:
classDiagram
direction TB
class Event["Event🔺"] {
}
class CourseInstance["CourseInstance🔺🟩"] {
}
class Course["Course🔺🟩"] {
syllabusSections
}
class new_LearningPath["new:LearningPath🔵"] {
Syllabus[] syllabusSections
}
class ListItem["ListItem🔺"] {
nextItem
}
class Syllabus["Syllabus🔺"] {
}
class new_LearningPathModule["new:LearningPathModule🔵"] {
ListItem[] itemListElement
LearningPathTopic nextItem
}
class LearningResource["LearningResource🔺"] {
}
class bio_TrainingMaterial["bio:TrainingMaterial🟩"] {
}
Course <|-- new_LearningPath
Course <|-- new_LearningPathModule
Syllabus <|-- new_LearningPathModule
ListItem <|-- new_LearningPathModule
LearningResource <|-- Course
LearningResource <|-- bio_TrainingMaterial
LearningResource <|-- Syllabus
Event <|-- CourseInstance
Concrete Example from Galaxy Training Network
The team mocked encoding the Introduction to Galaxy and Sequence analysis learning path on TeSS in this new schema. This learning path has the following structure:
- Module 1: Introduction to Galaxy
- A short introduction to Galaxy
- Galaxy Basics for genomics
- Module 2: Basics of Genome Sequence Analysis
- Quality Control
- Mapping
- An Introduction to Genome Assembly
- Chloroplast genome assembly
Here’s a mockup of how this could look in RDF:
@prefix dct: <http://purl.org/dc/terms/> .
@prefix ex: <http://example.org/> .
@prefix schema: <https://schema.org/> .
ex:GA_learning_path a schema:Course ;
dct:conformsTo <https://bioschemas.org/profiles/LearningPath> ;
schema:courseCode "GSA101" ;
schema:description "This learning path aims to teach you the basics of Galaxy and analysis of sequencing data. " ;
schema:name "Introduction to Galaxy and Sequence analysis" ;
schema:provider ex:ExampleUniversity ;
schema:syllabusSections ex:Module_1,
ex:Module_2 .
ex:Module_1 a schema:ItemList,
schema:ListItem,
schema:Syllabus ;
dct:conformsTo <https://bioschemas.org/profiles/LearningPathModule> ;
schema:itemListElement ex:TM11,
ex:TM12 ;
schema:name "Module 1: Introduction to Galaxy" ;
schema:nextItem ex:Module_2 ;
schema:teaches "Learn how to create a workflow" .
ex:TM11 a schema:LearningResource,
schema:ListItem ;
dct:conformsTo <https://bioschemas.org/profiles/TrainingMaterial> ;
schema:description "What is Galaxy" ;
schema:name "(1.1) A short introduction to Galaxy" ;
schema:nextItem ex:TM12 ;
schema:url "https://tess.elixir-europe.org/materials/hands-on-for-a-short-introduction-to-galaxy-tutorial?lp=1%3A1" .
Here’s the same thing from a graphical perspective:
graph TD
N1["Module 1: Introduction to Galaxy"]
N3["(1.2) Galaxy Basics for genomics"]
N1 -- itemListElement --> N3
N1["Module 1: Introduction to Galaxy"]
N2["(1.1) A short introduction to Galaxy"]
N1 -- itemListElement --> N2
N4["Module 2: Basics of Genome Sequence Analysis"]
N8["(2.4) Chloroplast genome assembly"]
N4 -- itemListElement --> N8
N2["(1.1) A short introduction to Galaxy"]
N3["(1.2) Galaxy Basics for genomics"]
N2 -- nextItem --> N3
N1["Module 1: Introduction to Galaxy"]
N4["Module 2: Basics of Genome Sequence Analysis"]
N1 -- nextItem --> N4
N7["(2.3) An Introduction to Genome Assembly"]
N8["(2.4) Chloroplast genome assembly"]
N7 -- nextItem --> N8
N4["Module 2: Basics of Genome Sequence Analysis"]
N5["(2.1) Quality Control"]
N4 -- itemListElement --> N5
N4["Module 2: Basics of Genome Sequence Analysis"]
N6["(2.2) Mapping"]
N4 -- itemListElement --> N6
N4["Module 2: Basics of Genome Sequence Analysis"]
N7["(2.3) An Introduction to Genome Assembly"]
N4 -- itemListElement --> N7
N6["(2.2) Mapping"]
N7["(2.3) An Introduction to Genome Assembly"]
N6 -- nextItem --> N7
N3["(1.2) Galaxy Basics for genomics"]
N5["(2.1) Quality Control"]
N3 -- nextItem --> N5
N5["(2.1) Quality Control"]
N6["(2.2) Mapping"]
N5 -- nextItem --> N6
Something that I became aware of while listening to discussions about learning path is the way that Schema.org models lists. I wonder why they don’t use the built-in RDF notions of lists and instead implemented their own formalism. I saw that this caused a lot of confusion for the team both during mocking and also during SPARQL querying.
I think the next steps in terms of learning paths is to create a concrete implementation in OERbsevatory - we have the benefit that the Python programming language provides a much more ergonomic abstraction over lists and collections. There’s a lot of content inside the Galaxy Training Network (GTN) that could be ingested into such a learning path. Towards this end, I gave a quick demo of Pydantic to the learning paths team and showed them how I typically go about data modeling.
I really enjoyed the BioHackathon, and in general, I am very happy to be attending more events to network with other academics in Germany. It was totally exhausting, too, which is why I didn’t manage to finish this in the week following the event.
In other open educational resource and learning materials news, we pre-printed the first ac academic article describing a specific use case for DALIA on arXiv in September: Teaching RDM in a smart advanced inorganic lab course and its provision in the DALIA platform. We’re currently finalizing a second article fully dedicated towards describing the DALIA platform which I hope can go on the arXiv in early January. Stay tuned!

