Mapping from SSSOM to Wikidata
At the
4th Ontologies4Chem Workshop
in Limburg an der Lahn, I proposed an initial crosswalk between the
Simple Standard for Sharing Ontological Mappings (SSSOM)
and the Wikidata semantic mapping data model. This
post describes the motivation for this proposal and the concrete implementation
I’ve developed in sssom-pydantic.
This work is part of the NFDI’s Ontology Harmonization and Mapping Working Group, which is interested in enabling interoperability between SSSOM and related data standards that encode semantic mappings.
The TL;DR for this post is that I implemented a mapping from SSSOM to Wikidata
in sssom-pydantic in
cthoyt/sssom-pydantic#32.
One high-level entrypoint is the following function, which reads an SSSOM file
and prepares
QuickStatements which can
be reviewed in the web browser, then uploaded to Wikidata.
This script can be run from Gist with
uv run https://gist.github.com/cthoyt/f38d37426a288989158a9804f74e731a#file-sssom-wikidata-demo-py
Semantic Mappings in SSSOM
The Simple Standard for Sharing Ontological Mappings (SSSOM) is a community-driven data standard for semantic mappings, which are necessary to support (semi-)automated data integration and knowledge integration, such as in the construction of knowledge graphs.
While SSSOM primary a tabular data format that is best serialized in TSV, it uses LinkML to formalize the semantics of each field such that SSSOM can be serialized to and read from OWL, RDF, and JSON-LD. Here’s a brief example:
| subject_id | subject_label | predicate_id | object_id | object_label | mapping_justification |
|---|---|---|---|---|---|
| wikidata:Q128700 | cell wall | skos:exactMatch | GO:0005618 | cell wall | semapv:ManualMappingCuration |
| wikidata:Q47512 | acetic acid | skos:exactMatch | CHEBI:15366 | acetic acid | semapv:ManualMappingCuration |
Semantic Mappings in Wikidata
Wikidata has two complementary formalisms for representing semantic mappings.
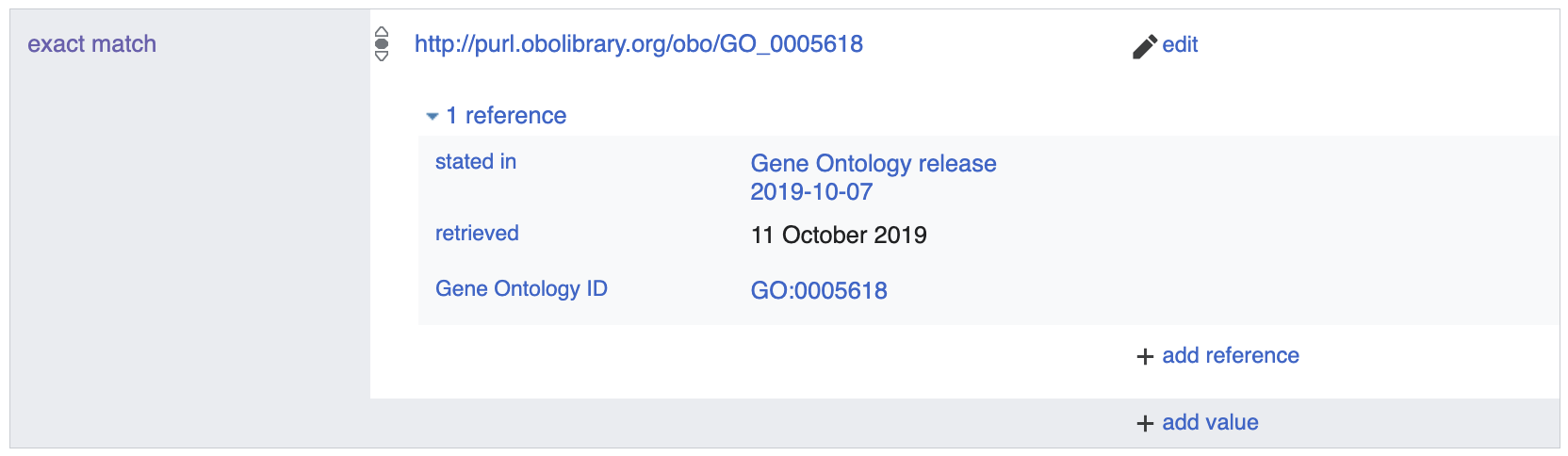
The first uses the
exact match (P2888) property
with a URI as the object. For example,
cell wall (Q128700) maps to the Gene
Ontology (GO) term for cell wall
by its URI http://purl.obolibrary.org/obo/GO_0005618.
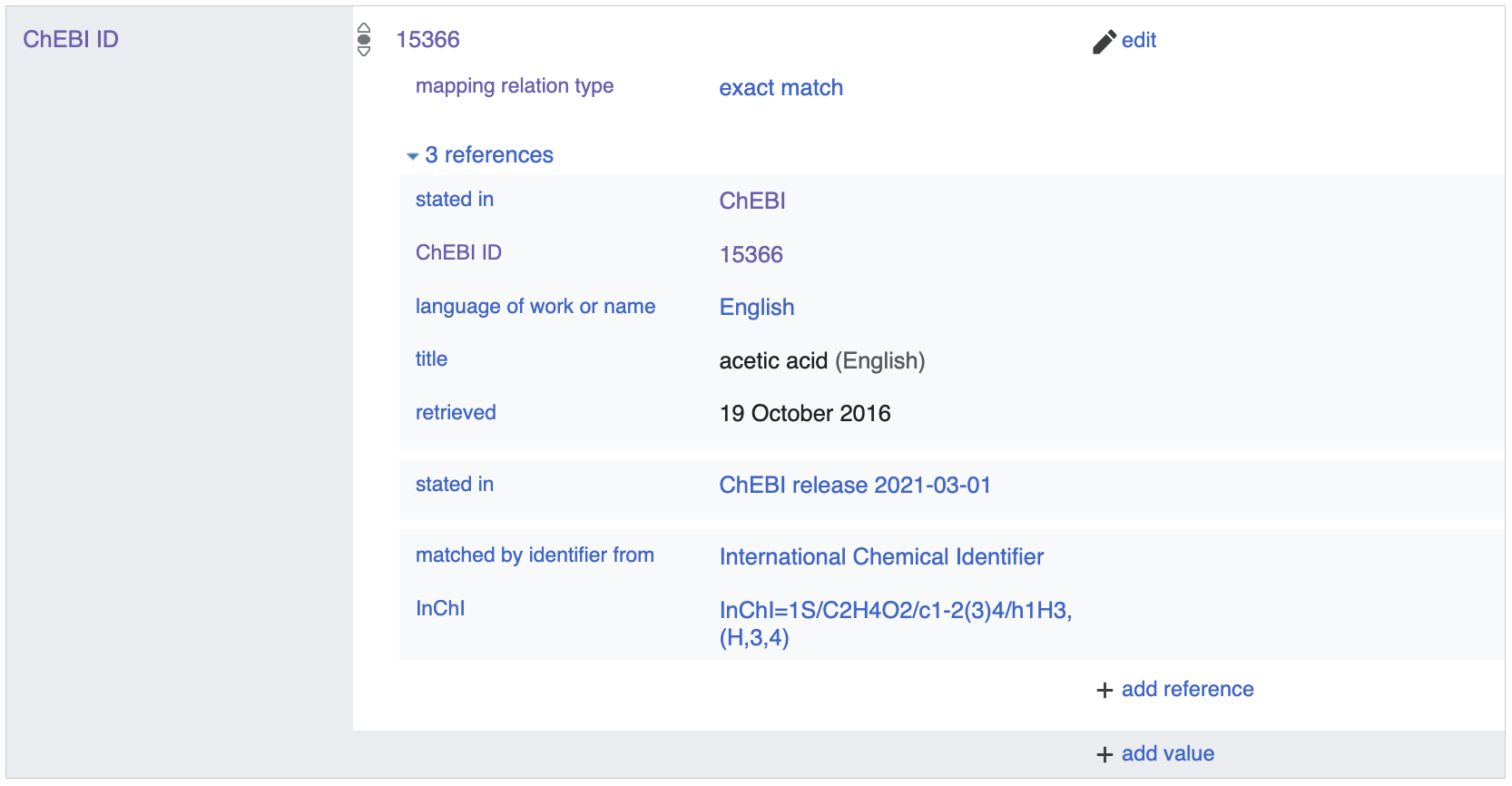
The second formalism uses semantic space-specific properties (e.g.
P683 for ChEBI) with local unique
identifiers as the object. For example,
acetic acid (Q47512) maps to the ChEBI
term for
acetic acid using
the P683 property for ChEBI and
local unique identifier for acetic acid (within ChEBI) 15366.
Wikidata has a data structure that enables annotating qualifiers onto triples. Therefore, other parts of semantic mappings modeled in SSSOM can be ported:
- Authors and reviewers can be mapped from ORCiD identifiers to Wikidata identifiers, then encoded using the S50 and S4032 properties, respectively
- A SKOS-flavored mapping predicate (i.e., exact, narrow, broad, close, related) can be encoded using the S4390 property
- The publication date can be encoded using the S577 property
- The license can be mapped from text to a Wikidata identifier, then encoded using the S275 property
Note that properties that normally start with a P when used in triples are
changed to start with an S when used as qualifiers. Other fields in SSSOM
could potentially be mapped to Wikidata later.
Finding Wikidata Properties using the Semantic Farm
The Semantic Farm (previously called the Bioregistry)
maintains mappings between prefixes that appear in compact URIs (CURIEs) and
their corresponding Wikidata properties. For example, the prefix
CHEBI maps to the Wikidata property
P683.

These mappings can be accessed in several ways:
- via the Semantic Farm’s SSSOM export. Note: this requires subsetting to mappings where Wikidata properties are the object.
- via the Semantic Farm’s live API,
-
via the Bioregistry Python package (this will get renamed to match Semantic Farm, eventually) using the following code:
import bioregistry # get bulk prefix_to_property = bioregistry.get_registry_map("wikidata") # get for a single resource resource = bioregistry.get_resource("chebi") chebi_wikidata_property_id = resource.get_mapped_prefix("wikidata")
Notable Implementation Details
I’ve previously built two package which were key to making this work:
wikidata-client, which interacts with the Wikidata SPARQL endpoint and has high-level wrappers around lookup functionality. I’m also aware of WikidataIntegrator - I’ve contributed several improvements, but working with its codebase doesn’t spark joy and the last time I tried to use it, it was fully broken due to some of its dependencies not working on modern Python.quickstatements-client, which implements an object model for QuickStatements v2 and an API client.
Along the way to this PR, I made improvements to the wikidata-client in cthoyt/wikidata-client#2 to add high-level functionality for looking up multiple Wikidata records based on values for a property (e.g., to support ORCID lookup in bulk).
All other changes were made in sssom-pydantic in
cthoyt/sssom-pydantic#32.
The other key challenge was to avoid adding duplicate information to Wikidata - unlike a simple triple store, we could accidentally end up with duplicate statements. Therefore, the sssom-pydantic implementation looks up all existing semantic mappings in Wikidata for entities appearing in an SSSOM file, then filters appropriately to avoid uploading duplicate mappings to Wikidata.
Pulling it All Together
This new module in sssom-pydantic implements the following interactive
workflows:
- Read an SSSOM file, convert mappings to Wikidata schema, then open a
QuickStatements tab in the web browser using
read_and_open_quickstatements() - Convert in-memory semantic mappings to the Wikidata schema, then open a
QuickStatements tab in the web browser using
open_quickstatements()
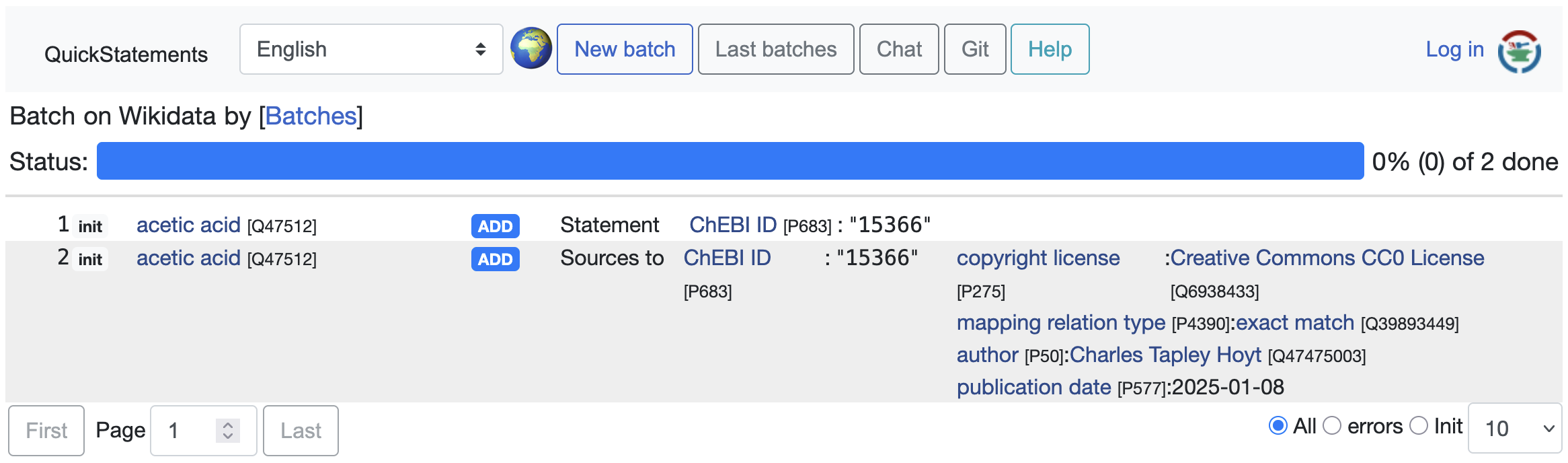
Here’s what the QuickStatements web interface looks like after preparing some demo mappings:
It also implements the following non-interactive workflows, which should be used with caution since they write directly to Wikidata:
- Read an SSSOM file, convert mappings to Wikidata schema, then post
non-interactively to Wikidata via QuickStatements using
read_and_post() - Convert in-memory semantic mappings to the Wikidata schema, then post
non-interactively to Wikidata via QuickStatements using
post()
I’m a bit hesitant to start uploading SSSOM content to Wikidata in bulk, because I don’t yet have a plan for how to maintain mappings that might change over time in their upstream single source of truth, e.g., mappings curated in Biomappings. Otherwise, I think this is a good proof of concept and would like to get feedback about additional qualifiers that could be added, and if the ones I chose so far were the best.