Mapping from SSSOM to JSKOS
JSKOS (JSON for Knowledge Organization Systems) is a JSON-based data model for representing terminologies, thesauri, classifications, and other semantic artifacts. Like the Simple Standard for Sharing Ontological Mappings (SSSOM), it can also encode semantic mappings. This post is about developing and implementing a crosswalk between them in the sssom-pydantic Python package.
Background on JSKOS
At its core, JSKOS implements the Simple Knowledge Organization System (SKOS) data model and extends it with a data model inspired by Wikidata with the following types:
JSKOS enables representing semantic mappings two ways:
- using the
narrower,broader, andrelatedslots in the Concept class that correspond to SKOS relationsskos:narrowMatch,skos:broadMatch, andskos:relatedMatch - using the
mappingsslot in the Concept class, which accepts a list of instances of the more generic Mapping class
Here’s how JSKOS represents an exact match from the Biomappings community curated mappings database between a Medical Subject Headings (MeSH) term and Chemical Entities of Biological Interest (ChEBI) ontology term for the chemical ammeline:
{
"license": [{ "uri": "https://spdx.org/licenses/CC0-1.0" }],
"uri": "https://w3id.org/biopragmatics/biomappings/sssom/biomappings.sssom.tsv",
"mappings": [
{
"type": ["http://www.w3.org/2004/02/skos/core#exactMatch"],
"subject_bundle": {
"member_set": [{ "uri": "http://id.nlm.nih.gov/mesh/C000089" }]
},
"object_bundle": {
"member_set": [{ "uri": "http://purl.obolibrary.org/obo/CHEBI_28646" }]
},
"justification": "https://w3id.org/semapv/vocab/ManualMappingCuration"
}
]
}
Notably, JSKOS is baked into the Cocoda mapping editor, which is being widely adopted in the humanities consortia of the NFDI.
Interoperability between SSSOM and JSKOS
Given the overlapping ability of the Simple Standard for Sharing Ontological Mappings (SSSOM) and JSKOS to represent semantic mappings, the JSKOS and SSSOM teams developed a crosswalk between JSKOS and SSSOM. Along the way, the SSSOM and JSKOS data models evolved to incorporate good ideas from the other, for example, the addition of a mapping identifier to SSSOM records to allow for referencing the SSSOM mapping itself.
The crosswalk is not (yet) lossless, for example, JSKOS does not yet have a mechanism to express information about lexical and other automated mappings. However, lossless conversion between data models isn’t always possible, nor is it always necessary, considering the different domains for which JSKOS and SSSOM were developed. That JSKOS was developed by researchers in the digital humanities and SSSOM was developed by researchers in the life and natural sciences can contextualize some of their discrepancies.
Technical Implementation
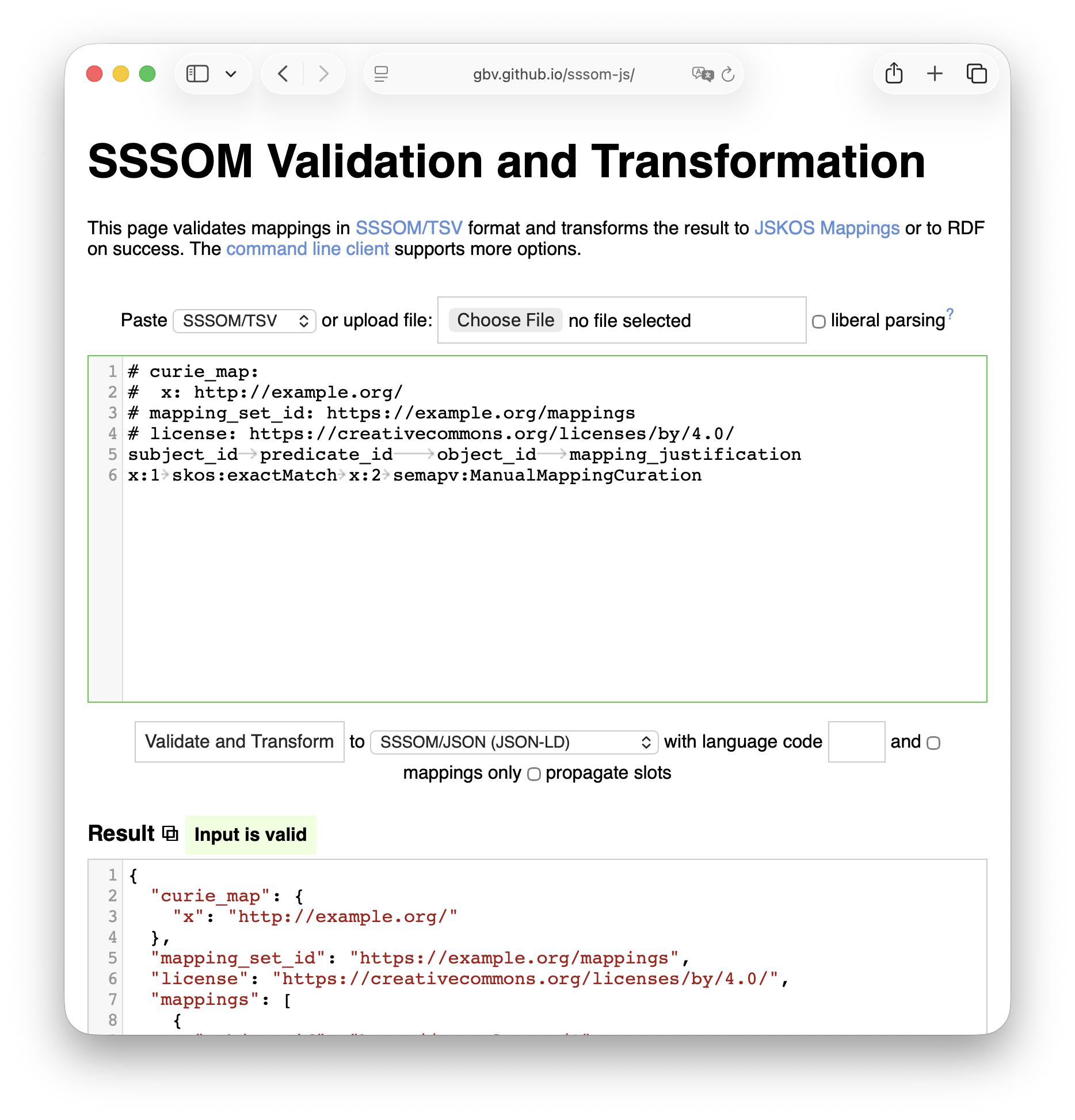
The sssom-js JavaScript package contains the first SSSOM to JSKOS converter and has an open issue for conversion back to SSSOM (TSV). It was developed by the JSKOS team, meaning that I have high confidence that the implemenation of the crosswalk is accurate.
While it can be invoked from the command line using npx like in
npx sssom-js --from tsv --to jskos --output output.json input.sssom.tsv, it
can also be explored in the first-party SSSOM Validation and Transformation
website.
Originally, my plan was to implement SSSOM to JSKOS export in the sssom-pydantic so it can be easily incorporated into other SSSOM-aware applications like SSSOM Curator and the Semantic Mapping Reasoner and Assembler.
I started by implementing an object model for JSKOS in Python using Pydantic in a dedicated package. This actually turned out to be very difficult to get to work in general because the JSKOS data model is hierarchical and does not always contain fields that make it possible to discriminate between which class a given arbitrary JSON object follows. This makes it difficult to use Pydantic’s nested discriminated unions feature, so I had to implement a custom solution.
Ultimately, I scrapped the idea of re-implementing the crosswalk myself (for
now) and instead defer to the sssom-js implementation (while wrapping it in an
idiomatic Python API). Once sssom-js implements a TSV exporter, I will have a
high-quality oracle against which to test my implementation. These first steps
were implemented in
cthoyt/sssom-pydantic#26.
Here’s what this looks like in Python:
import sssom_pydantic
from sssom_pydantic.contrib.jskos_export import to_jskos
url = "https://w3id.org/biopragmatics/biomappings/sssom/biomappings.sssom.tsv"
mappings, converter, metadata = sssom_pydantic.read(url)
jskos_concept = to_jskos(mappings, converter=converter, metadata=metadata)