Semantic Pydantic
Using Pydantic for encoding data models and FastAPI for implementing APIs on top of them has become a staple for many Python programmers. When this intersects with the semantic web, linked open data, and the natural sciences, we are still lacking a bridge to annotate our data models and APIs to make them more FAIR (findable, accessible, interoperable, and reusable). In this post, we build an extension to Pydantic and FastAPI to annotate data models’ fields and API endpoints’ query, path, and other parameters using the Bioregistry, a comprehensive catalog of metadata about semantic spaces from the semantic web and the natural sciences.
As a demonstration, we will build a data model and API that serves information about scholars.
First Steps with Pydantic
We’ll use Open Researcher and Contributor (ORCID) identifiers as primary keys, include the researcher’s name, and start with a single cross-reference, e.g., to the author’s DBLP identifier. We’ll encode this data model using Pydantic in the Python programming language as follows:
from pydantic import BaseModel, Field
class ScholarV1(BaseModel):
"""A model representing a researcher, who might have several IDs on different services."""
orcid: str = Field(...)
name: str = Field(...)
dblp: str | None = Field(None)
print(ScholarV1.schema_json(indent=2))
JSON Schema - Version 1
```json { "title": "ScholarV1", "description": "A model representing a researcher, who might have several IDs on different services.", "type": "object", "properties": { "orcid": { "title": "Orcid", "type": "string" }, "name": { "title": "Name", "type": "string" }, "dblp": { "title": "Dblp", "type": "string" } }, "required": ["orcid", "name"] } ```There are several places for improvement here:
- Correct capitalization of the titles (
ORCIDinstead ofOrcidandDBLPinstead ofDblp) - Add useful descriptions of what each field is
- Have regular expression patterns to validate input
- Give an example
All of these are possible to annotate into Pydantic’s Field object, but it
requires lots of effort and takes lots of space. Even worse, this might have to
be partially duplicated if multiple models share the same fields. In the example
below, I annotated ORCID but will skip the others for brevity.
from pydantic import BaseModel, Field
class ScholarV2(BaseModel):
"""A model representing a researcher, who might have several IDs on different services."""
orcid: str = Field(
...,
title="ORCID",
description="A stable, public identifier for a researcher from https://orcid.com",
pattern="^\d{4}-\d{4}-\d{4}-\d{3}(\d|X)$",
example="0000-0003-4423-4370",
)
name: str = Field(...)
dblp: str | None = Field(None)
print(ScholarV2.schema_json(indent=2))
JSON Schema - Version 2
```json { "title": "ScholarV2", "description": "A model representing a researcher, who might have several IDs on different services.", "type": "object", "properties": { "orcid": { "title": "ORCID", "description": "A stable, public identifier for a researcher from https://orcid.com", "pattern": "^\\d{4}-\\d{4}-\\d{4}-\\d{3}(\\d|X)$", "example": "0000-0003-4423-4370", "type": "string" }, "name": { "title": "Name", "type": "string" }, "dblp": { "title": "Dblp", "type": "string" } }, "required": ["orcid", "name"] } ```However, this was a lot of work. It would be nice if there were some database of all the semantic spaces in the semantic web and natural sciences that contained the name, description, regular expression pattern, and examples. Then, we could draw from this database to automatically populate our fields.
Adding Some Bioregistry Magic
The good news is that such a database exists - it’s called the Bioregistry. Each semantic space (e.g., ORCID, DBLP) gets a prefix which is usually an acronym for the name of the resource that serves as the primary key for the semantic space. These prefixes are also useful in making references to entities in the semantic space more FAIR (findable, accessible, interoperable, reusable) using the compact URI (CURIE) syntax, though this isn’t the goal of this demo.
I’ve mocked some Python code that bridges Pydantic and the Bioregistry in this repository (https://github.com/cthoyt/semantic-pydantic). I’m calling it Semantic Pydantic because it lets us annotate our data models with external metadata (and because it rhymes).
Here’s the same model as before, but now using a SemanticField that extends
Pydantic’s Field. It has a special keyword prefix that lets you give a
Bioregistry prefix, then it is smart enough to fill out all the fields on its
own. I also took the liberty of adding several more semantic spaces that
identify scholars like
Web of Science (wos),
Scopus, and even
GitHub.
from pydantic import BaseModel, Field
from semantic_pydantic import SemanticField
class ScholarV3(BaseModel):
"""A model representing a researcher, who might have several IDs on different services."""
orcid: str = SemanticField(..., prefix="orcid")
name: str = Field(..., example="Charles Tapley Hoyt")
wos: str | None = SemanticField(default=None, prefix="wos.researcher")
dblp: str | None = SemanticField(default=None, prefix="dblp.author")
github: str | None = SemanticField(default=None, prefix="github")
scopus: str | None = SemanticField(default=None, prefix="scopus")
semion: str | None = SemanticField(default=None, prefix="semion")
publons: str | None = SemanticField(default=None, prefix="publons.researcher")
authorea: str | None = SemanticField(default=None, prefix="authorea.author")
print(ScholarV3.schema_json(indent=2))
Finally, we can see a very detailed JSON schema, which includes everything from before plus additional context from the Bioregistry, including the prefix itself as well as mappings from the Bioregistry prefix to external registries like BARTOC, FAIRsharing, and others. Together, these make the data model more FAIR and support interoperability, since now it is possible to directly match the fields annotated with Bioregistry prefixes in this model to fields annotated with the same prefix in other models, even external to the project.
JSON Schema - Version 3
```json { "title": "ScholarV3", "description": "A model representing a researcher, who might have several IDs on different services.", "type": "object", "properties": { "orcid": { "title": "Open Researcher and Contributor", "description": "This field corresponds to a local unique identifier from Open Researcher and Contributor</a>.\n
Provenance
The semantics of this field are derived from the\n<a href=\"https://bioregistry.io/orcid\">orcid</a> entry in\nthe <a href=\"https://bioregistry.io\">Bioregistry</a>: a registry of semantic web and linked \nopen data compact URI (CURIE) prefixes and URI prefixes.\n
Description of Semantic Space
ORCID (Open Researcher and Contributor ID) is an open, non-profit, community-based effort to create and maintain a registry of unique identifiers for individual researchers. ORCID records hold non-sensitive information such as name, email, organization name, and research activities.", "pattern": "^\\d{4}-\\d{4}-\\d{4}-\\d{3}(\\d|X)$", "example": "0000-0003-4423-4370", "json_schema_extra": { "bioregistry": { "prefix": "orcid", "mappings": { "bartoc": "2021", "biocontext": "ORCID", "biolink": "ORCID", "fairsharing": "FAIRsharing.nx58jg", "go": "orcid", "miriam": "orcid", "n2t": "orcid", "wikidata": "P496" } } }, "type": "string" }, "name": { "title": "Name", "example": "Charles Tapley Hoyt", "type": "string" }, ... } } ``` Note that the extra semantic fields have been truncated for brevity. Each of them also has lots of detailed information.Web Application
Let’s take the next step to a web application using FastAPI. The goal of this web application will be to look up the information for a scholar in Wikidata based on their ORCID. You don’t really have to understand how the query works other than that it takes in an ORCID string and gives back an instance of the Scholar model we’ve been working on above.
The app uses annotations for the query parameters, path parameters, and other
inputs to routes using extensions of Pydantic Fields. So similar to before, we
can extend their custom fields to be semantic in Semantic Pydantic.
from fastapi import FastAPI
from semantic_pydantic import SemanticPath
app = FastAPI(title="Semantic Pydantic Demo")
Scholar = ... # defined before
@app.get("/api/orcid/{orcid}", response_model=Scholar)
def get_scholar_from_orcid(orcid: str = SemanticPath(prefix="orcid")):
"""Get xrefs for a researcher in Wikidata, given ORCID identifier."""
... # full implementation in https://github.com/cthoyt/semantic-pydantic
return Scholar(...)
The real power is how this translates to the API, and more importantly, the
automatically generated API documentation. First, the SemanticPath object
which we used in place of a normal fastapi.Path also knows it is for ORCID
identifiers. Second, the response model points to the Scholar class from before
which already knows about its semantics. Below, we see this in a screenshot of
the OpenAPI (formerly known as Swagger) user interface automatically generated
by FastAPI.
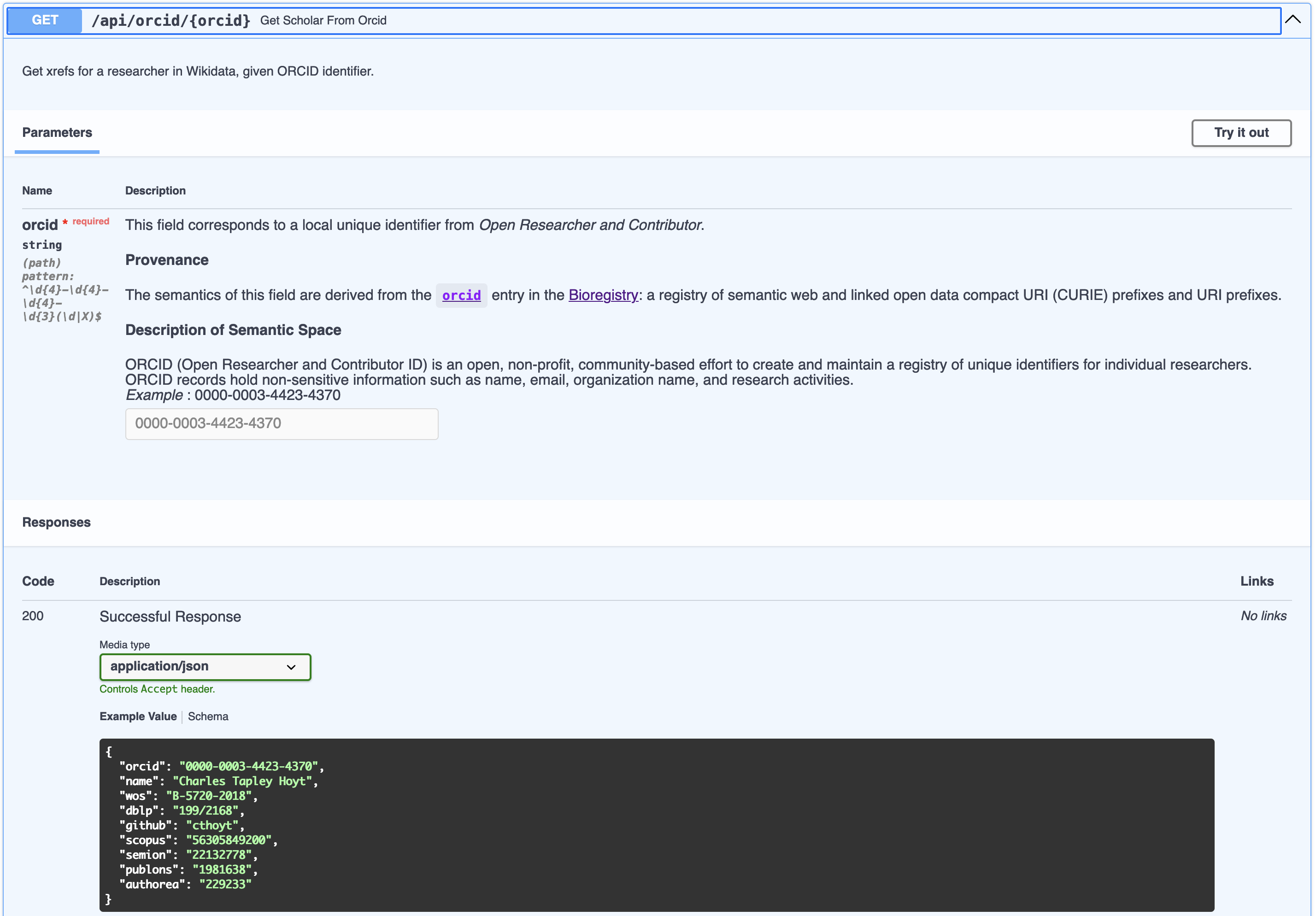
There are two big things to note here:
- All the detailed information about ORCID makes it into the parameter, also giving an example for users to get started
- All the detailed information from the response model (Scholar) gets shown as a real example.
Now, we have an API that is also annotated with detailed semantics. If you take a look at the OpenAPI JSON file, it has similar references to Bioregistry prefixes for the routes themselves, and directly reuses the JSON schema for the response model.
Next Steps
So far, this is a proof-of-concept that lives in an ad hoc repository. It’s not clear yet if this code is just a neat demo, whether it should live inside the Bioregistry Python package, I haven’t decided yet if this should go inside the Bioregistry Python package, or if it should be in a stand-alone package that might be extensible even further. There are a few other things to think about in the meantime:
Infrastructure for FAIR Models and APIs
The first version of this idea just throws the Bioregistry data into the JSON schema. It would be interesting to develop this infrastructure further, such as keeping a catalog of all APIs that consume or produce data models containing semantic fields. A few places this would be great:
- The
Cheminformatics Microservice
contains tons of references to
InChi Keys (
inchikey) - The Bioregistry could dog-food its own API
- The INDRA Discovery API could be refactored to use semantic inputs and outputs
There are also so many more examples, please let me know some services you think would benefit in the comments on my blog post. Looking forward, it’s also a question on how to automatically discover such semantic APIs (e.g., by cleverly searching GitHub) or if it would have to be a manually curated catalog.
Resolving URLs
A key feature of the Bioregistry is that it provides a way to take a local
unique identifier for an entity in a given semantic space and make a URL that
points to a web page describing the entity. For example, if you have an ORCID
identifier, you can make a URL for the ORCID page following the format
https://orcid.org/<put ID here>. It would be very cool to extend Semantic
Pydantic to add some properties that auto-generate URLs, like in the following:
from pydantic import BaseModel, Field
from semantic_pydantic import SemanticField
class Scholar(BaseModel):
orcid: str = SemanticField(..., prefix="orcid")
name: str = Field(...)
charlie = Scholar(orcid="0000-0003-4423-4370", name="Charles Tapley Hoyt")
assert charlie.orcid_url == 'https://orcid.org/0000-0003-4423-4370'
Run the Demo
The demo can be run by cloning the repository, installing its requirements, and
running the self-contained app.py.
git clone https://github.com/cthoyt/semantic-pydantic
cd semantic-pydantic
python -m pip install -r requirements.txt
python app.py
This is my first science post of 2024! I’m very happy that the Bioregistry is currently supported by the Chan Zuckerberg Initiative (CZI) under award 2023-329850.